Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
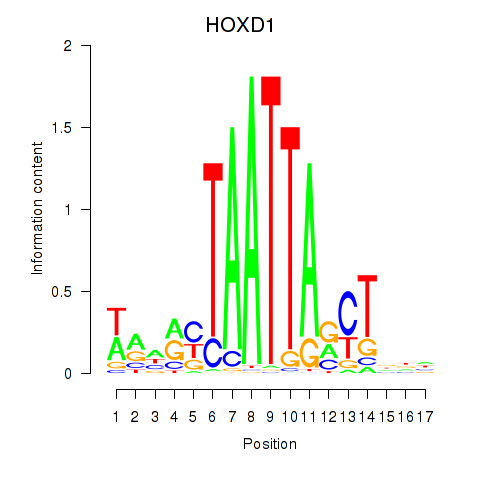
Results for HOXD1
Z-value: 0.37
Transcription factors associated with HOXD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD1
|
ENSG00000128645.11 | homeobox D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD1 | hg19_v2_chr2_+_177053307_177053402 | 0.29 | 5.8e-01 | Click! |
Activity profile of HOXD1 motif
Sorted Z-values of HOXD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_67535647 | 0.34 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_19458597 | 0.26 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr22_-_28490123 | 0.26 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr7_-_99716914 | 0.25 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr3_-_191000172 | 0.20 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr7_+_37723450 | 0.17 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr11_+_71900703 | 0.16 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_+_71900572 | 0.14 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr4_+_71019903 | 0.14 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr12_-_52761262 | 0.13 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr20_-_50418947 | 0.13 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr22_-_24951888 | 0.13 |
ENST00000404664.3
|
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr17_-_57229155 | 0.12 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_-_126327398 | 0.11 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr2_+_228736335 | 0.11 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chrX_+_107288239 | 0.11 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_71823796 | 0.11 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr5_+_148737562 | 0.10 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr15_-_55562479 | 0.10 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_64798826 | 0.10 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr4_+_144354644 | 0.10 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_-_71823715 | 0.10 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr2_+_191221240 | 0.10 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr7_-_55620433 | 0.09 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr7_+_99717230 | 0.09 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chrX_+_107288197 | 0.09 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr15_+_63050785 | 0.09 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr7_-_99717463 | 0.09 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr22_-_24951814 | 0.09 |
ENST00000407973.2
|
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr1_-_232651312 | 0.08 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr2_+_135596180 | 0.08 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_-_99716940 | 0.08 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_99716952 | 0.08 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrX_-_19504642 | 0.08 |
ENST00000469203.2
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr4_-_119759795 | 0.07 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr19_-_3557570 | 0.07 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr12_+_15699286 | 0.07 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr12_-_118797475 | 0.07 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_66300464 | 0.06 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_124329336 | 0.06 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr11_-_71823266 | 0.06 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr10_+_99205894 | 0.06 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr11_+_73498898 | 0.06 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr20_+_43990576 | 0.06 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr15_+_58702742 | 0.06 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr1_+_117544366 | 0.06 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr20_-_50419055 | 0.06 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr14_+_32798462 | 0.05 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_-_151431674 | 0.05 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr10_-_72142345 | 0.05 |
ENST00000373224.1
ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20
|
leucine rich repeat containing 20 |
| chr10_+_99205959 | 0.05 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr10_+_18549645 | 0.05 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr14_+_32798547 | 0.05 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chrX_+_43515467 | 0.05 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr1_-_151431647 | 0.05 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr5_+_66300446 | 0.04 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_+_18086392 | 0.04 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr19_-_36001113 | 0.04 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr11_-_26593649 | 0.04 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr12_+_29376592 | 0.04 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr1_-_151431909 | 0.04 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr4_-_150736962 | 0.04 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr12_+_29376673 | 0.03 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr2_+_135596106 | 0.03 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr4_-_74486347 | 0.03 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_-_92952433 | 0.03 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr15_-_55562582 | 0.03 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_109577202 | 0.03 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr14_+_56584414 | 0.02 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr15_-_37393406 | 0.01 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chrX_+_77154935 | 0.01 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr3_-_99569821 | 0.01 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr13_-_36050819 | 0.01 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr15_-_55563072 | 0.01 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_81399438 | 0.01 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr4_+_158493642 | 0.01 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr2_-_99871570 | 0.01 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr1_-_201140673 | 0.01 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr11_-_102709441 | 0.01 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr17_-_27418537 | 0.01 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr8_+_55528627 | 0.01 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr16_+_53133070 | 0.01 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_49833978 | 0.01 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr5_+_63802109 | 0.01 |
ENST00000334025.2
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein |
| chr5_-_20575959 | 0.01 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr1_-_190446759 | 0.00 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_139051839 | 0.00 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.4 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |