Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
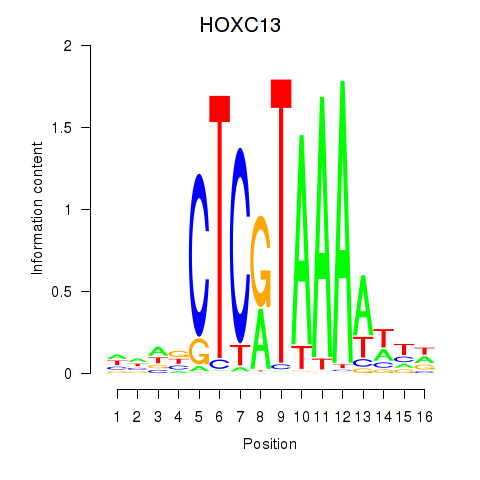
Results for HOXC13
Z-value: 0.98
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | homeobox C13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | -0.35 | 5.0e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_155533787 | 1.05 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr6_+_151358048 | 0.85 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr15_-_71184724 | 0.79 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr2_+_172309634 | 0.74 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chrM_+_8366 | 0.74 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr15_+_71185148 | 0.72 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr7_-_92777606 | 0.72 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr1_+_219347203 | 0.69 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr15_+_71228826 | 0.67 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_+_219347186 | 0.65 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr15_+_71184931 | 0.64 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chrM_+_8527 | 0.61 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr2_+_201390843 | 0.60 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr3_-_160117035 | 0.57 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr6_-_150217195 | 0.51 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr7_-_64023441 | 0.50 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr8_-_82608409 | 0.50 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr1_-_150208498 | 0.50 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_4647606 | 0.50 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr2_+_172778952 | 0.50 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr14_-_82089405 | 0.49 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr11_-_7904464 | 0.49 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr6_+_88182643 | 0.48 |
ENST00000369556.3
ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr4_+_141178440 | 0.47 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr11_-_104480019 | 0.46 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr12_+_102514019 | 0.46 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr14_-_50319482 | 0.46 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr1_+_222886694 | 0.46 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr1_-_150208412 | 0.46 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_123728548 | 0.45 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr9_-_32526299 | 0.44 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr2_+_161993465 | 0.43 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr16_-_66907139 | 0.41 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr21_+_25801041 | 0.41 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr1_-_150208363 | 0.40 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr14_-_88200641 | 0.40 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr1_-_89591749 | 0.39 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr5_-_39270725 | 0.39 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr14_-_50319758 | 0.38 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr1_-_150208320 | 0.38 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr2_-_37384175 | 0.38 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr5_+_138678131 | 0.37 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr14_-_80678512 | 0.37 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr4_+_128702969 | 0.36 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr2_+_161993412 | 0.36 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_64838712 | 0.35 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr6_-_11807277 | 0.35 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr19_-_23869999 | 0.34 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr4_-_128887069 | 0.34 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr2_+_162016827 | 0.33 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr4_+_119200215 | 0.33 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr9_+_40028620 | 0.32 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr7_+_64838786 | 0.32 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr3_-_98241760 | 0.31 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr7_-_32534850 | 0.31 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_+_174158732 | 0.31 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr1_+_174128639 | 0.31 |
ENST00000251507.4
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_-_92333873 | 0.30 |
ENST00000435962.2
|
TC2N
|
tandem C2 domains, nuclear |
| chrY_+_14958970 | 0.30 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_+_56471592 | 0.29 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr3_-_160116995 | 0.29 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr2_+_48010312 | 0.27 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr10_-_21186144 | 0.27 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr15_+_77713222 | 0.27 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr12_-_74796291 | 0.27 |
ENST00000551726.1
|
RP11-81H3.2
|
RP11-81H3.2 |
| chr6_+_134274322 | 0.27 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr14_+_96722539 | 0.27 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr11_+_107643129 | 0.27 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr2_+_33683109 | 0.27 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_150208291 | 0.26 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_86174065 | 0.26 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr6_+_134274354 | 0.26 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr2_+_162016916 | 0.26 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr12_+_67663056 | 0.26 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr21_+_47706537 | 0.25 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr2_-_223520770 | 0.25 |
ENST00000536361.1
|
FARSB
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr4_+_175204865 | 0.25 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr11_-_58378759 | 0.25 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr6_+_88117683 | 0.25 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr2_+_162016804 | 0.24 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_38186775 | 0.24 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr3_-_123339343 | 0.24 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr16_-_9770700 | 0.24 |
ENST00000561538.1
|
RP11-297M9.1
|
Uncharacterized protein |
| chr4_+_95679072 | 0.23 |
ENST00000515059.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_-_47407136 | 0.23 |
ENST00000462347.1
ENST00000371905.1 ENST00000310638.4 |
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr9_-_69229650 | 0.23 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr4_+_70894130 | 0.22 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr1_-_155880672 | 0.22 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr1_+_174128704 | 0.21 |
ENST00000457696.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr9_-_2844058 | 0.21 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr4_-_71532668 | 0.21 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr7_-_30066233 | 0.21 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr6_-_32784687 | 0.21 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr13_+_28813645 | 0.21 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr6_-_130031358 | 0.21 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr11_+_63449045 | 0.20 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chrX_+_133733457 | 0.20 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr9_-_5304432 | 0.20 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr15_+_77224045 | 0.20 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr1_+_90460661 | 0.20 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr19_+_52102540 | 0.20 |
ENST00000601315.1
|
AC018755.17
|
AC018755.17 |
| chr7_-_29152509 | 0.19 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr17_-_74639730 | 0.19 |
ENST00000589992.1
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr17_+_56769924 | 0.19 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr12_-_91546926 | 0.19 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_-_220220000 | 0.19 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr8_-_17555164 | 0.19 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr1_+_100435986 | 0.19 |
ENST00000532693.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr9_+_67977438 | 0.19 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr2_-_165424973 | 0.19 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr7_-_27169801 | 0.19 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr8_+_35649365 | 0.18 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr11_+_85566422 | 0.18 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr3_+_100428316 | 0.18 |
ENST00000479672.1
ENST00000476228.1 ENST00000463568.1 |
TFG
|
TRK-fused gene |
| chr2_+_138721850 | 0.18 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr10_+_94352956 | 0.18 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr19_+_21265028 | 0.18 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr6_+_56820152 | 0.17 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr1_+_213224572 | 0.17 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr1_+_158979680 | 0.17 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr7_-_152373216 | 0.16 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr7_-_16872932 | 0.16 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr3_+_99986036 | 0.16 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr3_+_147657764 | 0.16 |
ENST00000467198.1
ENST00000485006.1 |
RP11-71N10.1
|
RP11-71N10.1 |
| chr19_-_38085633 | 0.16 |
ENST00000593133.1
ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571
|
zinc finger protein 571 |
| chrX_+_119737806 | 0.15 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr3_+_50211240 | 0.15 |
ENST00000420831.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr5_-_58571935 | 0.15 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chrX_+_13707235 | 0.15 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr13_-_103451307 | 0.14 |
ENST00000376004.4
|
KDELC1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr12_+_96883347 | 0.14 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr13_+_103451399 | 0.14 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr19_+_21106081 | 0.14 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr1_+_246887349 | 0.14 |
ENST00000366510.3
|
SCCPDH
|
saccharopine dehydrogenase (putative) |
| chr15_+_69373184 | 0.14 |
ENST00000558147.1
ENST00000440444.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr6_+_21666633 | 0.14 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr14_+_37641012 | 0.13 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr1_-_145076068 | 0.13 |
ENST00000369345.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_158979686 | 0.13 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr2_+_33359687 | 0.13 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_174128536 | 0.13 |
ENST00000357444.6
ENST00000367689.3 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_+_73634537 | 0.12 |
ENST00000406768.1
|
PSEN1
|
presenilin 1 |
| chr19_+_16999654 | 0.12 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr7_+_23749894 | 0.12 |
ENST00000433467.2
|
STK31
|
serine/threonine kinase 31 |
| chr14_-_68000442 | 0.12 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr6_+_19837592 | 0.12 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr1_-_149832704 | 0.12 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr17_-_56769382 | 0.11 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr7_+_23749945 | 0.11 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr3_+_100428188 | 0.11 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr6_+_127898312 | 0.11 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr2_+_33359646 | 0.11 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_210444142 | 0.11 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr10_+_30723533 | 0.11 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_102953608 | 0.11 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr5_-_64920115 | 0.11 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr17_-_40170506 | 0.11 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr10_+_70847852 | 0.11 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr14_+_24702073 | 0.11 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr12_-_54689532 | 0.11 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr19_-_18632861 | 0.11 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr2_+_61372226 | 0.10 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr7_+_138915102 | 0.10 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr6_-_75953484 | 0.10 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr1_+_55107449 | 0.10 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr10_-_113943447 | 0.10 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr15_+_77223960 | 0.10 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr4_-_69536346 | 0.10 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr4_-_170533723 | 0.10 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr6_+_126221034 | 0.10 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr18_-_52989525 | 0.10 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr7_+_90012986 | 0.10 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr4_+_106631966 | 0.10 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr4_+_170581213 | 0.09 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr22_+_32439019 | 0.09 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr9_-_130533615 | 0.09 |
ENST00000373277.4
|
SH2D3C
|
SH2 domain containing 3C |
| chr2_-_122262593 | 0.09 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr4_+_190802670 | 0.09 |
ENST00000503609.1
|
RP11-463J17.1
|
RP11-463J17.1 |
| chr1_-_13331692 | 0.09 |
ENST00000353410.5
ENST00000376173.3 |
PRAMEF3
|
PRAME family member 3 |
| chr4_-_120243545 | 0.09 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr4_+_8321882 | 0.09 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr1_+_109656719 | 0.09 |
ENST00000457623.2
ENST00000529753.1 |
KIAA1324
|
KIAA1324 |
| chr3_-_179169181 | 0.09 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr5_+_169010638 | 0.09 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr10_-_70287172 | 0.09 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr12_-_13248598 | 0.09 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr17_+_43861680 | 0.09 |
ENST00000314537.5
|
CRHR1
|
corticotropin releasing hormone receptor 1 |
| chr12_+_45609862 | 0.08 |
ENST00000423947.3
ENST00000435642.1 |
ANO6
|
anoctamin 6 |
| chr17_-_10707410 | 0.08 |
ENST00000581851.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr7_+_23749767 | 0.08 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr7_+_63709496 | 0.08 |
ENST00000255746.4
|
ZNF679
|
zinc finger protein 679 |
| chr12_-_47473425 | 0.07 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr3_+_111630451 | 0.07 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr5_+_134181625 | 0.07 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chrX_-_135338503 | 0.07 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr2_+_210444298 | 0.07 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr3_-_49837254 | 0.07 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr1_+_55107489 | 0.07 |
ENST00000395690.2
|
MROH7
|
maestro heat-like repeat family member 7 |
| chr4_+_122722466 | 0.07 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr5_-_79287060 | 0.07 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.8 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.4 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.5 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 0.2 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 1.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.4 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 2.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of Cdc42 protein signal transduction(GO:0032489) NMDA glutamate receptor clustering(GO:0097114) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:1904796 | synaptic vesicle targeting(GO:0016080) regulation of core promoter binding(GO:1904796) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.4 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.3 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |