Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
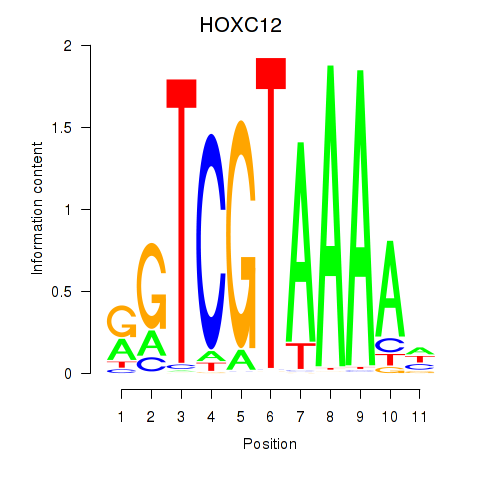
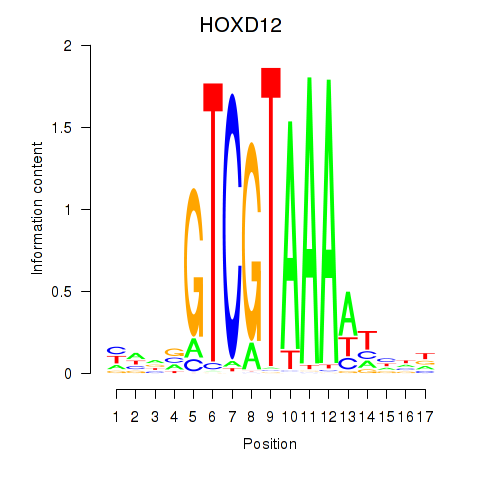
Results for HOXC12_HOXD12
Z-value: 0.63
Transcription factors associated with HOXC12_HOXD12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC12
|
ENSG00000123407.3 | homeobox C12 |
|
HOXD12
|
ENSG00000170178.5 | homeobox D12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD12 | hg19_v2_chr2_+_176964458_176964540 | 0.52 | 2.9e-01 | Click! |
| HOXC12 | hg19_v2_chr12_+_54348618_54348693 | 0.52 | 2.9e-01 | Click! |
Activity profile of HOXC12_HOXD12 motif
Sorted Z-values of HOXC12_HOXD12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_113354341 | 1.91 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr5_-_39270725 | 0.54 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr1_-_220219775 | 0.50 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr1_+_219347203 | 0.46 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr11_-_7904464 | 0.40 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr19_-_23869999 | 0.36 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr13_+_53030107 | 0.35 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr1_-_220220000 | 0.34 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr22_+_38035623 | 0.33 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr2_+_172778952 | 0.32 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr1_+_156308403 | 0.32 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr18_-_812517 | 0.32 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr5_+_158737824 | 0.31 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr16_+_9056563 | 0.31 |
ENST00000564485.1
|
RP11-77H9.8
|
RP11-77H9.8 |
| chr13_+_53029564 | 0.30 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr7_+_64838712 | 0.30 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr12_+_104337515 | 0.30 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr2_+_161993465 | 0.30 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_161993412 | 0.29 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_64838786 | 0.29 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr9_-_73029540 | 0.28 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr12_-_88535747 | 0.28 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr17_+_19091325 | 0.26 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr1_+_219347186 | 0.26 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr15_+_71185148 | 0.26 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_-_66524482 | 0.26 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr16_-_66907139 | 0.26 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr6_-_10694766 | 0.25 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr15_-_71184724 | 0.25 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr4_-_129491686 | 0.25 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr22_+_29168652 | 0.25 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr4_-_39367949 | 0.24 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr5_-_176433693 | 0.24 |
ENST00000507513.1
ENST00000511320.1 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr12_-_10007448 | 0.24 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr4_+_70894130 | 0.23 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr11_-_104480019 | 0.23 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr15_+_71184931 | 0.23 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_-_74964904 | 0.23 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr5_+_169011033 | 0.23 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr11_-_107328527 | 0.22 |
ENST00000282251.5
ENST00000433523.1 |
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr12_+_28299014 | 0.22 |
ENST00000538586.1
ENST00000536154.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr10_-_38146965 | 0.22 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr14_-_64846033 | 0.21 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr10_-_98347063 | 0.21 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr3_-_179322436 | 0.21 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr2_+_173955327 | 0.20 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr2_+_201450591 | 0.20 |
ENST00000374700.2
|
AOX1
|
aldehyde oxidase 1 |
| chr12_+_67663056 | 0.20 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr11_-_102668879 | 0.18 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr5_+_93954358 | 0.18 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr19_+_21265028 | 0.18 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr1_+_63989004 | 0.18 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr11_+_111896090 | 0.18 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr1_+_178694300 | 0.18 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr22_+_38035459 | 0.17 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr3_+_99979828 | 0.17 |
ENST00000485687.1
ENST00000344949.5 ENST00000394144.4 |
TBC1D23
|
TBC1 domain family, member 23 |
| chr1_-_246670614 | 0.17 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr15_-_79576156 | 0.17 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr11_+_127140956 | 0.17 |
ENST00000608214.1
|
RP11-480C22.1
|
RP11-480C22.1 |
| chr3_-_179322416 | 0.17 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr12_+_32832134 | 0.17 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr2_-_201753717 | 0.17 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr7_-_27169801 | 0.16 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr3_-_19975665 | 0.16 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr3_-_98241598 | 0.16 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr10_-_74856608 | 0.16 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr8_+_16884740 | 0.16 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr7_+_138915102 | 0.15 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr3_-_25706368 | 0.15 |
ENST00000424225.1
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa |
| chr11_-_3400442 | 0.15 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr1_+_149230680 | 0.15 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr5_+_138678131 | 0.15 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr14_+_31028348 | 0.15 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr3_+_179322573 | 0.14 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr14_-_50319482 | 0.14 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr8_+_33613265 | 0.14 |
ENST00000517292.1
|
RP11-317N12.1
|
RP11-317N12.1 |
| chr14_-_35591156 | 0.14 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr18_+_9103957 | 0.14 |
ENST00000400033.1
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr4_-_111120334 | 0.14 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr3_+_179322481 | 0.14 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_158975744 | 0.14 |
ENST00000426592.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr12_-_95945246 | 0.14 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr1_-_101491319 | 0.14 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr2_+_67624430 | 0.13 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr12_+_32832203 | 0.13 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr2_+_234526272 | 0.13 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr12_+_123259063 | 0.13 |
ENST00000392441.4
ENST00000539171.1 |
CCDC62
|
coiled-coil domain containing 62 |
| chr3_+_32737027 | 0.13 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr15_+_79166065 | 0.12 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr12_-_13248598 | 0.12 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr17_-_79359154 | 0.12 |
ENST00000572077.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr2_+_55746746 | 0.12 |
ENST00000406691.3
ENST00000349456.4 ENST00000407816.3 ENST00000403007.3 |
CCDC104
|
coiled-coil domain containing 104 |
| chr14_-_50319758 | 0.12 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr6_+_64281906 | 0.12 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr17_+_56769924 | 0.12 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr5_-_65018834 | 0.12 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_+_111896320 | 0.12 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr21_+_17214724 | 0.12 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr9_-_124976185 | 0.12 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr4_-_156298087 | 0.12 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr1_+_199996702 | 0.12 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_+_100818009 | 0.12 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr12_+_104697504 | 0.12 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr20_-_44540686 | 0.11 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr1_+_199996733 | 0.11 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr22_+_22723969 | 0.11 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr7_+_94536514 | 0.11 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr15_-_55657428 | 0.11 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr8_-_66701319 | 0.11 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr15_+_72410629 | 0.11 |
ENST00000340912.4
ENST00000544171.1 |
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr3_-_131221790 | 0.11 |
ENST00000512877.1
ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr7_-_112758665 | 0.11 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr10_-_1071796 | 0.10 |
ENST00000277517.1
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr14_-_22005197 | 0.10 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_+_71122974 | 0.10 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr13_+_52586517 | 0.10 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr5_-_176433582 | 0.10 |
ENST00000506128.1
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr4_+_110354928 | 0.10 |
ENST00000504968.2
ENST00000399100.2 ENST00000265175.5 |
SEC24B
|
SEC24 family member B |
| chr4_+_26585686 | 0.10 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr6_+_108616243 | 0.10 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr12_-_110939870 | 0.10 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr3_-_195310802 | 0.10 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr13_+_78315528 | 0.10 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_+_234545092 | 0.10 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_37571845 | 0.10 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr15_+_66797455 | 0.10 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr4_+_48833312 | 0.09 |
ENST00000508293.1
ENST00000513391.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr4_-_141348999 | 0.09 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr19_-_23941680 | 0.09 |
ENST00000402377.3
|
ZNF681
|
zinc finger protein 681 |
| chr12_+_28605426 | 0.09 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr8_-_82608409 | 0.09 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr19_+_21579908 | 0.09 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr12_+_10658201 | 0.09 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr13_+_34392200 | 0.09 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr4_-_70080449 | 0.09 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr5_+_93954039 | 0.09 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr8_+_121925301 | 0.09 |
ENST00000517739.1
|
RP11-369K17.1
|
RP11-369K17.1 |
| chr1_-_26231589 | 0.09 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr12_-_30848914 | 0.09 |
ENST00000256079.4
|
IPO8
|
importin 8 |
| chr15_+_66797627 | 0.09 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr19_+_7413835 | 0.09 |
ENST00000576789.1
|
CTB-133G6.1
|
CTB-133G6.1 |
| chr5_+_56471592 | 0.09 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr9_-_88896977 | 0.09 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr4_-_129208030 | 0.09 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr9_+_125703282 | 0.09 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr4_-_111120132 | 0.09 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_+_31608054 | 0.09 |
ENST00000320985.10
ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr13_-_79233314 | 0.08 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr3_-_165555200 | 0.08 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr2_+_101869262 | 0.08 |
ENST00000289382.3
|
CNOT11
|
CCR4-NOT transcription complex, subunit 11 |
| chr6_-_656963 | 0.08 |
ENST00000380907.2
|
HUS1B
|
HUS1 checkpoint homolog b (S. pombe) |
| chr4_-_129207942 | 0.08 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr2_-_136594740 | 0.08 |
ENST00000264162.2
|
LCT
|
lactase |
| chr7_-_112758589 | 0.08 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr5_+_145718587 | 0.08 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr4_-_111119804 | 0.08 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr8_-_80942061 | 0.08 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr13_-_29293058 | 0.08 |
ENST00000380814.4
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr6_-_28554977 | 0.08 |
ENST00000452236.2
|
SCAND3
|
SCAN domain containing 3 |
| chr3_+_138327417 | 0.08 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_+_107738240 | 0.08 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr2_+_61372226 | 0.08 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr14_+_24702073 | 0.08 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr19_+_42212526 | 0.08 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr4_+_122722466 | 0.08 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr5_+_140767452 | 0.08 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr3_+_132036207 | 0.08 |
ENST00000336375.5
ENST00000495911.1 |
ACPP
|
acid phosphatase, prostate |
| chr6_-_106773491 | 0.08 |
ENST00000360666.4
|
ATG5
|
autophagy related 5 |
| chr10_+_12238171 | 0.08 |
ENST00000378900.2
ENST00000442050.1 |
CDC123
|
cell division cycle 123 |
| chr17_+_34890807 | 0.08 |
ENST00000429467.2
ENST00000592983.1 |
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr17_+_71228793 | 0.08 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr3_-_196911002 | 0.08 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr14_+_24702127 | 0.08 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr10_+_6625733 | 0.07 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr12_+_90313394 | 0.07 |
ENST00000549551.1
|
RP11-654D12.2
|
RP11-654D12.2 |
| chr17_+_71228740 | 0.07 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr9_-_77502636 | 0.07 |
ENST00000449912.2
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr12_-_15114191 | 0.07 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr3_+_137906154 | 0.07 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr17_-_191188 | 0.07 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr11_+_36616044 | 0.07 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr4_+_48833234 | 0.07 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr7_+_138145145 | 0.07 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr13_+_113030658 | 0.07 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr6_-_110011718 | 0.07 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr13_+_34392185 | 0.07 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr3_+_138327542 | 0.07 |
ENST00000360570.3
ENST00000393035.2 |
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_+_24701819 | 0.07 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_+_156308245 | 0.07 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr4_-_114682224 | 0.06 |
ENST00000342666.5
ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr12_-_15114658 | 0.06 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_-_201753980 | 0.06 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr10_-_12238071 | 0.06 |
ENST00000491614.1
ENST00000537776.1 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chrX_-_133792480 | 0.06 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr3_+_143690640 | 0.06 |
ENST00000315691.3
|
C3orf58
|
chromosome 3 open reading frame 58 |
| chr13_+_49551020 | 0.06 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr4_-_69536346 | 0.06 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr7_+_154720173 | 0.06 |
ENST00000397551.2
|
PAXIP1-AS2
|
PAXIP1 antisense RNA 2 |
| chr17_-_26127525 | 0.06 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr21_-_46359760 | 0.06 |
ENST00000330551.3
ENST00000397841.1 ENST00000380070.4 |
C21orf67
|
chromosome 21 open reading frame 67 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC12_HOXD12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.3 | GO:1900244 | synaptic vesicle recycling via endosome(GO:0036466) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0061565 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 2.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of Cdc42 protein signal transduction(GO:0032489) NMDA glutamate receptor clustering(GO:0097114) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.2 | 1.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.3 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0034617 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |