Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
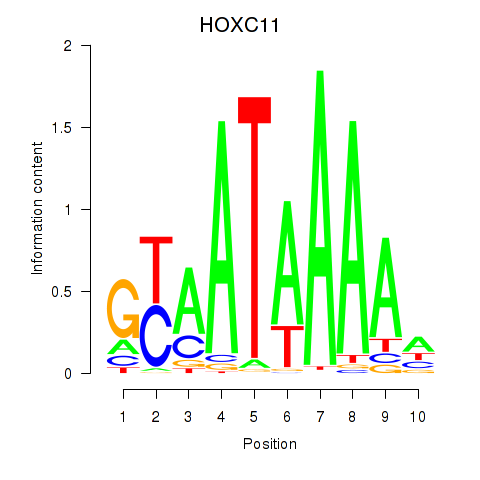
Results for HOXC11
Z-value: 0.78
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | homeobox C11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg19_v2_chr12_+_54366894_54366922 | 0.46 | 3.6e-01 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26156551 | 0.95 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr7_-_20826504 | 0.67 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr12_+_54384370 | 0.64 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr17_+_79369249 | 0.42 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr2_+_175199674 | 0.40 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr2_-_190044480 | 0.40 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr19_+_35634146 | 0.37 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr1_+_62439037 | 0.34 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr5_-_157161727 | 0.32 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr5_-_159766528 | 0.29 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr6_+_32812568 | 0.28 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_-_73793975 | 0.25 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr3_-_20053741 | 0.25 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr16_-_21863541 | 0.23 |
ENST00000543654.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr13_-_46626820 | 0.23 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr1_+_209878182 | 0.23 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_+_27440229 | 0.23 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr18_+_68002675 | 0.22 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr9_+_97766409 | 0.21 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr19_+_4402659 | 0.19 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr3_+_15045419 | 0.19 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr15_+_76352178 | 0.19 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr1_+_12524965 | 0.19 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_-_10504285 | 0.18 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr2_+_10223908 | 0.18 |
ENST00000425235.1
|
AC104794.4
|
AC104794.4 |
| chr14_-_25479811 | 0.18 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr9_+_105757590 | 0.18 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr11_-_47521309 | 0.17 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr7_+_128784712 | 0.17 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr19_-_19626351 | 0.17 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr7_+_134464376 | 0.17 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr1_-_12679171 | 0.16 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr7_-_47578840 | 0.16 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr5_+_176731572 | 0.16 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr22_+_41697520 | 0.16 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr8_+_26435915 | 0.16 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr17_-_77924627 | 0.16 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr11_-_65359947 | 0.16 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr7_-_47579188 | 0.16 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr18_+_72922710 | 0.16 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr16_-_67978016 | 0.15 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr15_+_67418047 | 0.15 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr14_+_97925151 | 0.15 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr10_+_47658234 | 0.15 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr17_-_2117600 | 0.14 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr1_-_12677714 | 0.14 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr10_+_47894023 | 0.14 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr14_-_69444957 | 0.14 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr4_+_160188889 | 0.13 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr20_+_42984330 | 0.13 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr12_+_54410664 | 0.13 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr9_-_133814527 | 0.13 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr19_+_19626531 | 0.13 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr9_+_97766469 | 0.13 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr15_+_36887069 | 0.13 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr7_-_122840015 | 0.13 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr6_+_15246501 | 0.13 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr9_+_82188077 | 0.12 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_-_19626838 | 0.12 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr18_-_74728998 | 0.12 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr7_+_22766766 | 0.12 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr12_-_28124903 | 0.12 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr10_+_114710516 | 0.11 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr16_+_15031300 | 0.11 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr21_+_35736302 | 0.11 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chrX_-_107682702 | 0.11 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr5_+_145718587 | 0.11 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr22_+_39916558 | 0.11 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr17_-_38928414 | 0.11 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr19_-_45927622 | 0.11 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chrX_+_107683096 | 0.11 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr16_-_21663950 | 0.11 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr22_+_40573921 | 0.11 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr5_-_36301984 | 0.11 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr16_-_2314222 | 0.11 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr22_-_30642782 | 0.11 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr18_-_52989217 | 0.10 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr5_+_148521454 | 0.10 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr17_-_39684550 | 0.10 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr7_-_41742697 | 0.10 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr12_-_123201337 | 0.10 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr11_+_63606373 | 0.10 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr6_-_41703952 | 0.10 |
ENST00000358871.2
ENST00000403298.4 |
TFEB
|
transcription factor EB |
| chr9_+_82187487 | 0.10 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_+_55996316 | 0.10 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr10_-_73497581 | 0.09 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr10_-_96829246 | 0.09 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr14_+_73706308 | 0.09 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr10_-_13523073 | 0.09 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr2_+_166428839 | 0.09 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_+_64001962 | 0.09 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr5_+_159656437 | 0.09 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr20_+_58713514 | 0.09 |
ENST00000432910.1
|
RP5-1043L13.1
|
RP5-1043L13.1 |
| chr8_-_109799793 | 0.09 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr8_+_134203273 | 0.09 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr17_+_7533439 | 0.09 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr12_-_54071181 | 0.09 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chrX_+_105192423 | 0.09 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr6_+_12290586 | 0.08 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr8_+_134203303 | 0.08 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr16_+_14805546 | 0.08 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr13_-_81801115 | 0.08 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr9_+_82187630 | 0.08 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_-_98189055 | 0.08 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr12_-_111358372 | 0.08 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr12_+_70219052 | 0.08 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr11_-_27723158 | 0.08 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr16_+_57653989 | 0.07 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr4_+_95972822 | 0.07 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_112443767 | 0.07 |
ENST00000550233.1
ENST00000546962.1 ENST00000550800.2 |
TMEM116
|
transmembrane protein 116 |
| chrX_+_14547632 | 0.07 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr17_+_7211656 | 0.07 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr1_+_151739131 | 0.07 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chrX_-_133792480 | 0.07 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr13_+_109248500 | 0.07 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr19_-_39924349 | 0.07 |
ENST00000602153.1
|
RPS16
|
ribosomal protein S16 |
| chr17_-_46690839 | 0.07 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr7_+_141418962 | 0.07 |
ENST00000493845.1
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr12_-_95510743 | 0.07 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr12_+_25205568 | 0.07 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr16_+_67906919 | 0.06 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr13_-_46626847 | 0.06 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr17_-_79876010 | 0.06 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr2_+_208423891 | 0.06 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr12_+_1800179 | 0.06 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr18_-_12656715 | 0.06 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr17_-_46657473 | 0.06 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr1_+_23345930 | 0.06 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr11_-_62473776 | 0.05 |
ENST00000278893.7
ENST00000407022.3 ENST00000421906.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr6_+_88054530 | 0.05 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr1_+_23345943 | 0.05 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr19_+_49468558 | 0.05 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr1_-_94147385 | 0.05 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_87794150 | 0.05 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr17_+_79027155 | 0.05 |
ENST00000572918.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr17_-_38821373 | 0.05 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr2_+_158114051 | 0.05 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr9_-_133814455 | 0.05 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr12_-_11339543 | 0.05 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr16_-_21431078 | 0.04 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr16_+_57653625 | 0.04 |
ENST00000567553.1
ENST00000565314.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr12_+_25205666 | 0.04 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr6_+_152011628 | 0.04 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr14_-_80782219 | 0.04 |
ENST00000553594.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr18_-_48351743 | 0.04 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr1_+_229440129 | 0.04 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr9_+_95736758 | 0.04 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr8_+_104831440 | 0.04 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr5_+_148521381 | 0.04 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr7_+_141811539 | 0.04 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr4_-_48683188 | 0.04 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr5_-_115872142 | 0.04 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_+_41236325 | 0.03 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr7_+_114055052 | 0.03 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr17_-_46806540 | 0.03 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr7_-_81399438 | 0.03 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_+_89156799 | 0.03 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr5_-_115872124 | 0.03 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr18_-_53019208 | 0.03 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr8_-_59412717 | 0.03 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr8_-_114449112 | 0.03 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr2_+_210443993 | 0.03 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr15_-_83952071 | 0.03 |
ENST00000569704.1
|
BNC1
|
basonuclin 1 |
| chr19_+_36036477 | 0.03 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr1_-_204135450 | 0.03 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr19_-_10420459 | 0.02 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr8_+_104831472 | 0.02 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_74726710 | 0.02 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr9_+_130026756 | 0.02 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_-_37216055 | 0.02 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_68872954 | 0.02 |
ENST00000394342.2
|
PROKR1
|
prokineticin receptor 1 |
| chr11_-_62473706 | 0.02 |
ENST00000403550.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr13_-_46679185 | 0.02 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr9_+_12693336 | 0.02 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chrX_-_19817869 | 0.02 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr11_-_62473684 | 0.02 |
ENST00000448568.2
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr7_-_81399355 | 0.02 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr9_-_73736511 | 0.02 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr4_+_169633310 | 0.02 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_215179188 | 0.02 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr1_-_200379129 | 0.01 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr20_+_58179582 | 0.01 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_-_27205136 | 0.01 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr11_+_34663913 | 0.01 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr2_-_157198860 | 0.01 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr16_+_57653854 | 0.01 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chrX_+_99899180 | 0.01 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr9_-_124976154 | 0.01 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr12_+_80838126 | 0.01 |
ENST00000266688.5
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q |
| chr12_-_52779433 | 0.01 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr2_-_50574856 | 0.01 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr15_-_68497657 | 0.01 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr2_+_211421262 | 0.01 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr9_-_124989804 | 0.01 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr7_-_11871815 | 0.01 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr5_-_34043310 | 0.01 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr4_-_74088800 | 0.01 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr1_+_75600567 | 0.01 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr12_-_71148413 | 0.01 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr2_+_210444298 | 0.01 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |