Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
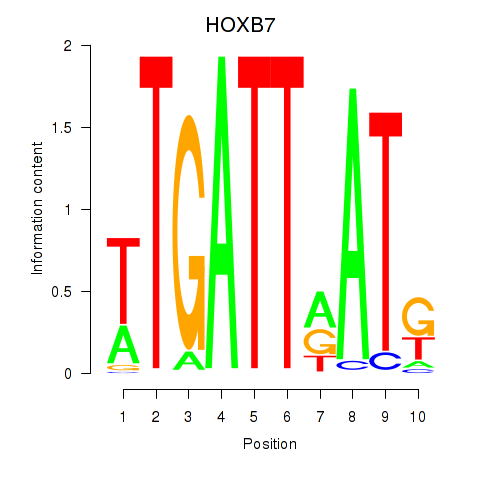
Results for HOXB7
Z-value: 0.44
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | 0.26 | 6.1e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_38557561 | 0.28 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr12_+_52056548 | 0.28 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr12_-_86650077 | 0.27 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr14_+_77425972 | 0.27 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr12_+_43086018 | 0.27 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr11_-_111649074 | 0.23 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr16_-_21289627 | 0.23 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr3_-_46608010 | 0.21 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr12_+_22852791 | 0.21 |
ENST00000413794.2
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr14_-_74417096 | 0.19 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr16_+_29127282 | 0.19 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr2_-_228028829 | 0.18 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr1_-_168513229 | 0.18 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2 |
| chr15_+_76352178 | 0.18 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr12_+_21525818 | 0.18 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr3_+_141105705 | 0.17 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_-_203735484 | 0.17 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr16_+_21623958 | 0.16 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr7_+_135777671 | 0.16 |
ENST00000445293.2
ENST00000435996.1 |
AC009784.3
|
AC009784.3 |
| chr3_+_52570610 | 0.15 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr3_-_4927447 | 0.15 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr6_+_116782527 | 0.15 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr4_+_71600063 | 0.15 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_+_124103625 | 0.15 |
ENST00000594963.1
|
AL161784.1
|
Uncharacterized protein |
| chr11_-_111649015 | 0.15 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr8_-_130799134 | 0.15 |
ENST00000276708.4
|
GSDMC
|
gasdermin C |
| chr15_+_83098710 | 0.14 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chr3_+_141105235 | 0.14 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr18_+_22040593 | 0.14 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr2_-_99917639 | 0.14 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr10_-_21786179 | 0.14 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr8_+_9953061 | 0.14 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr5_+_140227357 | 0.13 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr15_+_84904525 | 0.13 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr7_-_77325545 | 0.13 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chr16_+_66442411 | 0.13 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr19_+_49199209 | 0.13 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr3_-_196242233 | 0.13 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr12_-_67197760 | 0.13 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr4_+_139694701 | 0.13 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr1_+_12524965 | 0.12 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_+_48189612 | 0.12 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr17_+_7758374 | 0.12 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr3_+_158787041 | 0.12 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr3_-_71114066 | 0.12 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr4_+_169013666 | 0.12 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr10_+_24755416 | 0.12 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr1_-_179112189 | 0.12 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr18_+_20714525 | 0.12 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr1_-_179112173 | 0.11 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr6_-_116833500 | 0.11 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr7_+_75024903 | 0.11 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr20_+_36531544 | 0.11 |
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr4_+_144354644 | 0.11 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_-_155011483 | 0.11 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr8_+_39770803 | 0.11 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr13_-_110438914 | 0.11 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr20_+_36531499 | 0.11 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr11_-_58378759 | 0.11 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr8_+_98900132 | 0.11 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr16_+_22501658 | 0.11 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr21_+_43619796 | 0.11 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr7_-_7782204 | 0.11 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr22_+_46449674 | 0.10 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr1_+_152178320 | 0.10 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr3_+_111393501 | 0.10 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr6_-_2634820 | 0.10 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr19_-_57656570 | 0.10 |
ENST00000269834.1
|
ZIM3
|
zinc finger, imprinted 3 |
| chr17_-_57229155 | 0.10 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr19_-_33360647 | 0.10 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr3_+_187930429 | 0.10 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr15_+_85923856 | 0.10 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr10_+_17270214 | 0.10 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr2_+_27719697 | 0.10 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr10_+_47894572 | 0.10 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr8_+_9953214 | 0.10 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr15_+_82722225 | 0.10 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr15_-_28957469 | 0.10 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr15_-_82641706 | 0.10 |
ENST00000439287.4
|
GOLGA6L10
|
golgin A6 family-like 10 |
| chr9_-_85882145 | 0.10 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr15_+_85923797 | 0.09 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr16_-_84220633 | 0.09 |
ENST00000566732.1
ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr15_-_37393406 | 0.09 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr2_-_109605663 | 0.09 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr1_+_117963209 | 0.09 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr3_+_159943362 | 0.09 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr5_-_127418573 | 0.09 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr19_+_48774586 | 0.09 |
ENST00000594024.1
ENST00000595408.1 ENST00000315849.1 |
ZNF114
|
zinc finger protein 114 |
| chr18_+_21464737 | 0.09 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr11_-_126870655 | 0.09 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr14_+_74417192 | 0.09 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr11_-_126870683 | 0.09 |
ENST00000525704.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr19_+_38880252 | 0.09 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr17_-_46703826 | 0.09 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr16_-_28634874 | 0.09 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_+_21168630 | 0.08 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr4_-_102267953 | 0.08 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr17_+_76494911 | 0.08 |
ENST00000598378.1
|
DNAH17-AS1
|
DNAH17 antisense RNA 1 |
| chr4_+_113739244 | 0.08 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr10_-_98273668 | 0.08 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr8_-_67874805 | 0.08 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr12_-_95510743 | 0.08 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr19_+_19030497 | 0.08 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr4_+_158493642 | 0.08 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr15_+_86098670 | 0.08 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr9_+_105757590 | 0.08 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr22_-_22337146 | 0.08 |
ENST00000398793.2
|
TOP3B
|
topoisomerase (DNA) III beta |
| chr1_+_168148273 | 0.08 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr13_+_42614176 | 0.08 |
ENST00000540693.1
|
DGKH
|
diacylglycerol kinase, eta |
| chr16_-_66764119 | 0.08 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr7_-_22862406 | 0.08 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr5_-_78809950 | 0.08 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr3_+_111393659 | 0.08 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr9_-_128246769 | 0.08 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr16_-_24942411 | 0.08 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr17_+_45973516 | 0.08 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr16_-_24942273 | 0.08 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr4_+_71600144 | 0.08 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr8_-_67090825 | 0.08 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr5_-_10761206 | 0.08 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr19_-_51920952 | 0.08 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr6_-_110736742 | 0.08 |
ENST00000368924.3
ENST00000368923.3 |
DDO
|
D-aspartate oxidase |
| chr17_+_8191815 | 0.07 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr4_-_120243545 | 0.07 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr16_+_12059091 | 0.07 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_+_43477455 | 0.07 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr12_-_92821922 | 0.07 |
ENST00000538965.1
ENST00000378487.2 |
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand |
| chr2_-_208030295 | 0.07 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_+_20187174 | 0.07 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr1_+_244227632 | 0.07 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr13_-_33760216 | 0.07 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr9_-_75488984 | 0.07 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr5_+_98109322 | 0.07 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr6_+_144904334 | 0.07 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr17_+_58018269 | 0.07 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr19_+_19030478 | 0.07 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr8_-_91618285 | 0.07 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr2_-_230787879 | 0.07 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr6_-_15548591 | 0.07 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr10_-_47239738 | 0.07 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr22_+_40573921 | 0.07 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr6_+_42584847 | 0.07 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr21_+_41029235 | 0.07 |
ENST00000380618.1
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr1_-_146068184 | 0.07 |
ENST00000604894.1
ENST00000369323.3 ENST00000479926.2 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr3_+_159557637 | 0.06 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_+_47894023 | 0.06 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr6_+_28317685 | 0.06 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr20_-_50179368 | 0.06 |
ENST00000609943.1
ENST00000609507.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr14_-_24584138 | 0.06 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr19_+_52693259 | 0.06 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr11_+_107650219 | 0.06 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr16_+_16434185 | 0.06 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chrX_-_102757802 | 0.06 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr5_-_33297946 | 0.06 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr14_-_74959978 | 0.06 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chr10_+_81967456 | 0.06 |
ENST00000422847.1
|
LINC00857
|
long intergenic non-protein coding RNA 857 |
| chr5_+_125759140 | 0.06 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr7_+_141695671 | 0.06 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr11_-_8795787 | 0.06 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr12_-_11339543 | 0.06 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr15_-_75748115 | 0.06 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_-_43266680 | 0.06 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr19_-_48753064 | 0.06 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr1_-_157069590 | 0.06 |
ENST00000454449.2
|
ETV3L
|
ets variant 3-like |
| chr12_-_118628350 | 0.06 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr4_-_111536534 | 0.06 |
ENST00000503456.1
ENST00000513690.1 |
RP11-380D23.2
|
RP11-380D23.2 |
| chr8_-_116681123 | 0.06 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr9_+_2029019 | 0.06 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_78359999 | 0.05 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chrX_+_57618269 | 0.05 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr12_-_71031185 | 0.05 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chrX_+_120181457 | 0.05 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr16_-_84220604 | 0.05 |
ENST00000567759.1
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr6_-_44281043 | 0.05 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr18_+_44526744 | 0.05 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr12_+_133066137 | 0.05 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr11_+_44117260 | 0.05 |
ENST00000358681.4
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr11_+_44117099 | 0.05 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr10_+_69869237 | 0.05 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr16_+_15489603 | 0.05 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr15_+_67418047 | 0.05 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr11_-_123525289 | 0.05 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr8_-_124749609 | 0.05 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr1_+_75600567 | 0.05 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr10_-_17243579 | 0.05 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr11_-_104769141 | 0.05 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr3_+_140947563 | 0.05 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr21_+_44313375 | 0.05 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr7_+_116654935 | 0.05 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr14_-_65769392 | 0.05 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr2_-_197226875 | 0.05 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr19_+_20959098 | 0.05 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr7_+_2636522 | 0.05 |
ENST00000423196.1
|
IQCE
|
IQ motif containing E |
| chr19_+_41698927 | 0.05 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr1_-_28241024 | 0.05 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chrX_+_135388147 | 0.05 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr12_-_65146717 | 0.05 |
ENST00000545273.1
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr10_-_15902449 | 0.05 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr9_+_128509624 | 0.05 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr14_+_51955831 | 0.05 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) cellular response to cocaine(GO:0071314) positive regulation of fear response(GO:1903367) regulation of corticosterone secretion(GO:2000852) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.0 | GO:2000981 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.0 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.0 | GO:0052362 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.0 | GO:0008892 | guanine deaminase activity(GO:0008892) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |