Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HOXB5
Z-value: 0.39
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.5 | homeobox B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg19_v2_chr17_-_46671323_46671323 | 0.07 | 9.0e-01 | Click! |
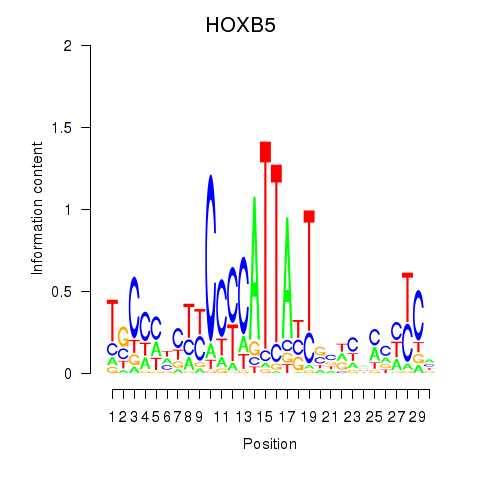
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_102704783 | 0.33 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chr1_+_146373546 | 0.16 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr1_+_154229547 | 0.15 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr17_-_17140436 | 0.13 |
ENST00000285071.4
ENST00000389169.5 ENST00000417064.1 |
FLCN
|
folliculin |
| chr1_+_113263199 | 0.12 |
ENST00000361886.3
|
FAM19A3
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
| chr7_-_102252589 | 0.12 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chrX_+_47077632 | 0.11 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr7_+_125557914 | 0.11 |
ENST00000411856.1
|
AC005276.1
|
AC005276.1 |
| chr6_-_31509506 | 0.10 |
ENST00000449757.1
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr3_+_190281229 | 0.10 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr19_-_16045220 | 0.10 |
ENST00000326742.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr8_-_133097902 | 0.09 |
ENST00000262283.5
|
OC90
|
Otoconin-90 |
| chr20_-_6034672 | 0.09 |
ENST00000378858.4
|
LRRN4
|
leucine rich repeat neuronal 4 |
| chr16_-_84651647 | 0.09 |
ENST00000564057.1
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr5_+_133842243 | 0.08 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr12_-_53614155 | 0.08 |
ENST00000543726.1
|
RARG
|
retinoic acid receptor, gamma |
| chr12_-_53614043 | 0.08 |
ENST00000338561.5
|
RARG
|
retinoic acid receptor, gamma |
| chr19_-_42724261 | 0.07 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr16_-_58004992 | 0.07 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr1_+_162351503 | 0.07 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr12_-_6798616 | 0.06 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr6_-_97285336 | 0.06 |
ENST00000229955.3
ENST00000417980.1 |
GPR63
|
G protein-coupled receptor 63 |
| chr17_+_46970127 | 0.06 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_-_70409953 | 0.06 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr16_-_28936007 | 0.06 |
ENST00000568703.1
ENST00000567483.1 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr17_+_46970178 | 0.06 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_-_6798410 | 0.06 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr1_+_155579979 | 0.06 |
ENST00000452804.2
ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_+_41157421 | 0.06 |
ENST00000372654.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr15_-_72523454 | 0.06 |
ENST00000565154.1
ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM
|
pyruvate kinase, muscle |
| chr1_+_157963391 | 0.06 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr16_-_1821721 | 0.05 |
ENST00000219302.3
|
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr6_-_31509714 | 0.05 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr3_-_127541194 | 0.05 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr20_-_48184638 | 0.05 |
ENST00000244043.4
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase |
| chr12_+_53693466 | 0.05 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr11_-_57089774 | 0.05 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr12_+_56915713 | 0.05 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr12_-_6798523 | 0.05 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr19_+_34745442 | 0.04 |
ENST00000299505.6
ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355
|
KIAA0355 |
| chr9_-_130700080 | 0.04 |
ENST00000373110.4
|
DPM2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr5_+_175298573 | 0.04 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr16_-_30381580 | 0.04 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr1_-_119530428 | 0.03 |
ENST00000369429.3
|
TBX15
|
T-box 15 |
| chr10_-_103603568 | 0.03 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr2_-_220173685 | 0.03 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr1_+_41157361 | 0.03 |
ENST00000427410.2
ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr6_-_31510181 | 0.03 |
ENST00000458640.1
ENST00000396172.1 ENST00000417556.2 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr13_-_64650144 | 0.03 |
ENST00000456627.1
|
LINC00355
|
long intergenic non-protein coding RNA 355 |
| chr19_-_50143452 | 0.03 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr15_+_41913690 | 0.03 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr4_-_68620053 | 0.03 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr10_-_103603523 | 0.03 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr20_+_44074071 | 0.03 |
ENST00000595203.1
|
AL031663.2
|
CDNA FLJ26875 fis, clone PRS08969; Uncharacterized protein |
| chr6_-_133119649 | 0.02 |
ENST00000367918.1
|
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr17_+_46970134 | 0.02 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_+_56915776 | 0.02 |
ENST00000550726.1
ENST00000542360.1 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr14_-_24020858 | 0.02 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr1_+_116915855 | 0.02 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr7_+_120969045 | 0.02 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr3_+_187420101 | 0.02 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr11_-_57089671 | 0.02 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr3_-_164914640 | 0.02 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr15_-_75017711 | 0.02 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr12_+_57984943 | 0.02 |
ENST00000422156.3
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chrX_+_100333709 | 0.01 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr12_-_56101647 | 0.01 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chrX_-_107225600 | 0.01 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr6_+_167525277 | 0.01 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr15_+_74218787 | 0.01 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr4_-_114900831 | 0.01 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chrX_+_48380205 | 0.01 |
ENST00000446158.1
ENST00000414061.1 |
EBP
|
emopamil binding protein (sterol isomerase) |
| chr6_-_133119668 | 0.01 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr1_+_154975110 | 0.01 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr16_-_1821496 | 0.01 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr7_-_102252038 | 0.00 |
ENST00000461209.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr14_+_21945335 | 0.00 |
ENST00000262709.3
ENST00000457430.2 ENST00000448790.2 |
TOX4
|
TOX high mobility group box family member 4 |
| chr16_-_9770700 | 0.00 |
ENST00000561538.1
|
RP11-297M9.1
|
Uncharacterized protein |
| chr18_+_32558208 | 0.00 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr8_+_91233750 | 0.00 |
ENST00000523406.1
|
LINC00534
|
long intergenic non-protein coding RNA 534 |
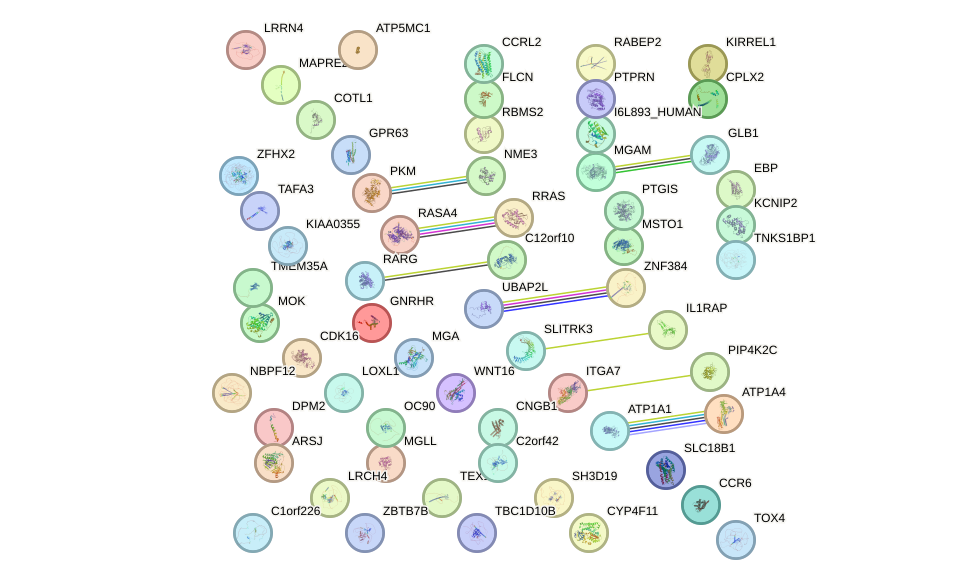
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |