Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
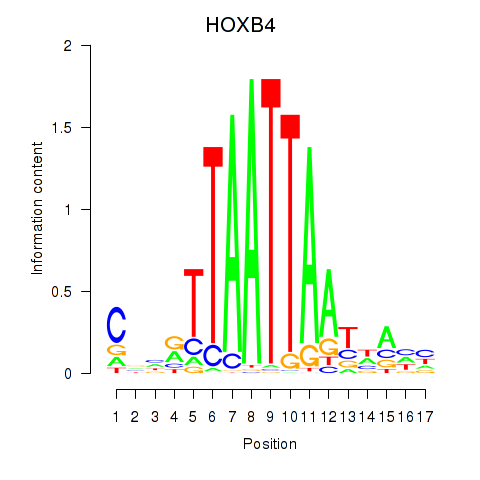
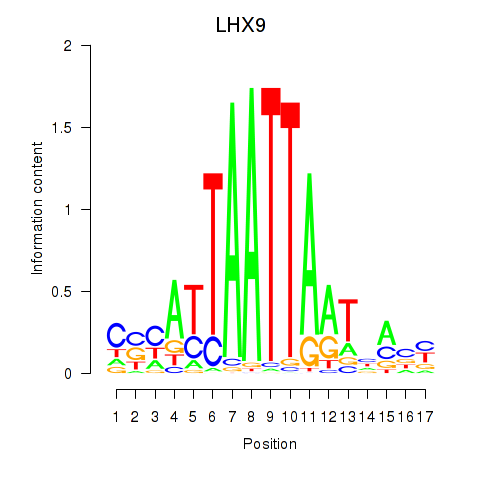
Results for HOXB4_LHX9
Z-value: 0.60
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB4
|
ENSG00000182742.5 | homeobox B4 |
|
LHX9
|
ENSG00000143355.11 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX9 | hg19_v2_chr1_+_197881592_197881635 | 0.71 | 1.2e-01 | Click! |
| HOXB4 | hg19_v2_chr17_-_46657473_46657473 | -0.30 | 5.7e-01 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_20187174 | 0.75 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr14_-_45252031 | 0.38 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr11_-_63376013 | 0.38 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr4_-_39033963 | 0.37 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr19_+_54466179 | 0.36 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr17_-_45266542 | 0.35 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr4_-_120243545 | 0.34 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr14_-_51027838 | 0.34 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_82957433 | 0.32 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr4_+_80584903 | 0.31 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr6_-_38670897 | 0.31 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr11_+_65265141 | 0.30 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr8_-_90996459 | 0.29 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr6_-_33663474 | 0.27 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr16_+_15489629 | 0.27 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr14_+_55494323 | 0.26 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr4_-_185275104 | 0.26 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr6_+_130339710 | 0.26 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_+_28390637 | 0.25 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr9_+_22646189 | 0.25 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chrX_+_108779004 | 0.25 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr8_-_90996837 | 0.25 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr14_+_31046959 | 0.24 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr4_+_95128748 | 0.24 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_-_38821373 | 0.23 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chrX_+_72783026 | 0.23 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr8_-_93978216 | 0.23 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_-_10022735 | 0.23 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_10978957 | 0.23 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chrX_+_84258832 | 0.22 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr4_-_103749105 | 0.21 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_67831576 | 0.21 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr1_+_225600404 | 0.20 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr6_+_26440700 | 0.20 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr14_+_62164340 | 0.20 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr4_-_103749313 | 0.19 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_+_136070614 | 0.19 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr12_+_34175398 | 0.19 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr12_-_8803128 | 0.19 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr5_+_129083772 | 0.19 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr4_+_76481258 | 0.18 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr17_+_57233087 | 0.18 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr3_+_44840679 | 0.18 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr14_+_55493920 | 0.18 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr3_+_23851928 | 0.18 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr12_+_147052 | 0.17 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr1_-_197115818 | 0.17 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr4_+_141264597 | 0.17 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr5_+_40841410 | 0.17 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr12_+_64798095 | 0.16 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_+_75970672 | 0.16 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chrX_-_71458802 | 0.16 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr18_-_32870148 | 0.16 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr11_+_118398178 | 0.16 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr5_+_89770696 | 0.16 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr9_-_99540328 | 0.16 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr4_+_158493642 | 0.16 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr4_+_25162253 | 0.15 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr9_-_95640218 | 0.15 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr5_-_159546396 | 0.15 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr6_-_109702885 | 0.14 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr18_+_20494078 | 0.14 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr1_-_212965104 | 0.14 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr5_+_112849373 | 0.14 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr2_+_29001711 | 0.13 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr15_+_90735145 | 0.13 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr14_-_39639523 | 0.13 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chrX_+_150565653 | 0.13 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_+_212965170 | 0.13 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr15_-_55489097 | 0.13 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chrX_+_77166172 | 0.13 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr3_+_186692745 | 0.13 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr4_-_170948361 | 0.13 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr8_-_93978333 | 0.13 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr9_-_115095229 | 0.12 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr16_-_14109841 | 0.12 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr19_+_36239576 | 0.12 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr2_+_103089756 | 0.12 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr4_+_26324474 | 0.11 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_+_89770664 | 0.11 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr8_-_93978357 | 0.11 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr11_-_107729287 | 0.11 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_+_136649311 | 0.11 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr6_-_116833500 | 0.11 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr15_-_60771280 | 0.11 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr2_+_187454749 | 0.11 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr11_+_5710919 | 0.11 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr8_-_93978346 | 0.11 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr2_-_27886676 | 0.10 |
ENST00000337768.5
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chrM_+_10758 | 0.10 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr9_-_77643307 | 0.10 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr2_+_102953608 | 0.10 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr8_-_30670384 | 0.10 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr12_+_20963647 | 0.10 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr14_-_35591679 | 0.10 |
ENST00000557278.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr14_+_45366472 | 0.10 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr19_+_50016610 | 0.10 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_+_48823896 | 0.10 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr3_+_38537960 | 0.10 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr5_-_102898465 | 0.10 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr12_+_20963632 | 0.10 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr5_-_36152031 | 0.09 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr10_+_35894338 | 0.09 |
ENST00000321660.1
|
GJD4
|
gap junction protein, delta 4, 40.1kDa |
| chr16_-_1538765 | 0.09 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chr10_-_27529486 | 0.09 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_-_103748880 | 0.09 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_-_93978309 | 0.09 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr2_+_172778952 | 0.09 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr13_+_78315295 | 0.09 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_-_140201333 | 0.09 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr11_+_12766583 | 0.09 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr14_-_35591433 | 0.09 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr2_-_161056762 | 0.09 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr18_-_10701979 | 0.09 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr4_+_37455536 | 0.08 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr5_-_70316737 | 0.08 |
ENST00000194097.4
|
NAIP
|
NLR family, apoptosis inhibitory protein |
| chr10_+_99627889 | 0.08 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr6_+_64345698 | 0.08 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr12_-_74686314 | 0.08 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr2_+_30670077 | 0.08 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr9_-_26947220 | 0.08 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr17_+_56833184 | 0.08 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr1_-_242612726 | 0.08 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr2_+_234637754 | 0.08 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_196743912 | 0.08 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr14_+_35591858 | 0.08 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chr7_+_23145884 | 0.08 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr4_-_36245561 | 0.08 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr14_+_24641062 | 0.08 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chrX_-_21776281 | 0.08 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr17_-_57229155 | 0.08 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_+_152214098 | 0.08 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr7_+_133812052 | 0.08 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr11_-_124670550 | 0.08 |
ENST00000239614.4
|
MSANTD2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr4_-_39979576 | 0.08 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr2_+_26624775 | 0.08 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr5_+_151338426 | 0.07 |
ENST00000524034.1
|
CTB-12O2.1
|
CTB-12O2.1 |
| chr7_+_141438118 | 0.07 |
ENST00000265304.6
ENST00000498107.1 ENST00000467681.1 ENST00000465582.1 ENST00000463093.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr2_+_237994519 | 0.07 |
ENST00000392008.2
ENST00000409334.1 ENST00000409629.1 |
COPS8
|
COP9 signalosome subunit 8 |
| chrX_-_102510045 | 0.07 |
ENST00000360000.4
|
TCEAL8
|
transcription elongation factor A (SII)-like 8 |
| chr20_+_5731083 | 0.07 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr12_+_32687221 | 0.07 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr4_+_119809984 | 0.07 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr4_+_11470867 | 0.07 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr8_-_112248400 | 0.07 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr2_-_191115229 | 0.07 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr14_+_50234309 | 0.07 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr11_-_13011081 | 0.07 |
ENST00000532541.1
ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958
|
long intergenic non-protein coding RNA 958 |
| chr2_+_219125714 | 0.07 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr10_-_99030395 | 0.07 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chrX_-_102510120 | 0.07 |
ENST00000451678.1
|
TCEAL8
|
transcription elongation factor A (SII)-like 8 |
| chr1_+_152178320 | 0.07 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr14_-_80697396 | 0.07 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chrX_-_102510126 | 0.07 |
ENST00000372685.3
|
TCEAL8
|
transcription elongation factor A (SII)-like 8 |
| chr14_+_35591735 | 0.07 |
ENST00000604948.1
ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391
|
KIAA0391 |
| chr19_-_58326267 | 0.07 |
ENST00000391701.1
|
ZNF552
|
zinc finger protein 552 |
| chr3_-_98619999 | 0.07 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr2_-_228244013 | 0.06 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr17_+_14277419 | 0.06 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chr1_-_77685084 | 0.06 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr14_-_53258314 | 0.06 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr2_-_161056802 | 0.06 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr7_+_151791095 | 0.06 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr12_+_81110684 | 0.06 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr5_-_119669160 | 0.06 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr16_+_24549014 | 0.06 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr21_-_15583165 | 0.06 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr4_-_74486217 | 0.06 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_103997862 | 0.06 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr17_+_67498538 | 0.06 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr7_+_102553430 | 0.06 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr10_+_64280207 | 0.06 |
ENST00000395251.1
|
ZNF365
|
zinc finger protein 365 |
| chrX_+_86772707 | 0.06 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr7_+_138145076 | 0.06 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr13_+_36050881 | 0.06 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr13_-_31038370 | 0.06 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr7_+_138915102 | 0.06 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr14_-_35591156 | 0.06 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr3_-_141747950 | 0.06 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_-_86849883 | 0.06 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr7_+_116654935 | 0.05 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr16_-_66583701 | 0.05 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr4_+_157997273 | 0.05 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr3_-_160823040 | 0.05 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr13_-_24007815 | 0.05 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr2_-_70520539 | 0.05 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr14_+_35591928 | 0.05 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr5_-_89770582 | 0.05 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr19_-_36001113 | 0.05 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr14_+_35591020 | 0.05 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr10_+_90660832 | 0.05 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr19_-_3772209 | 0.05 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr11_-_327537 | 0.05 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr8_-_101571933 | 0.05 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr8_-_90769422 | 0.05 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr2_+_182850551 | 0.05 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_27886330 | 0.05 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chrX_+_100663243 | 0.05 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr7_-_108168580 | 0.05 |
ENST00000453085.1
|
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr16_+_15489603 | 0.05 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |