Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
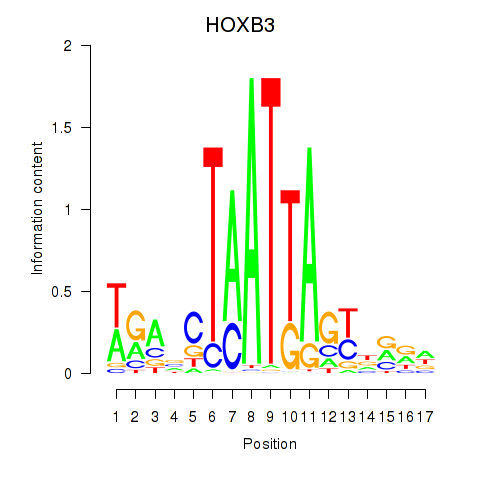
Results for HOXB3
Z-value: 0.48
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | homeobox B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg19_v2_chr17_-_46667594_46667619 | 0.58 | 2.3e-01 | Click! |
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_99716914 | 0.38 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_26045603 | 0.29 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr1_+_62439037 | 0.26 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr15_-_44969086 | 0.24 |
ENST00000434130.1
ENST00000560780.1 |
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr3_-_48659193 | 0.20 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr11_-_60010556 | 0.19 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr17_+_74846230 | 0.18 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr17_-_40337470 | 0.18 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr12_+_78359999 | 0.17 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr2_+_191221240 | 0.17 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr19_+_9296279 | 0.16 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr1_+_81106951 | 0.15 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr5_+_148737562 | 0.15 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr19_+_50031547 | 0.15 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_-_92777606 | 0.15 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_-_102651343 | 0.14 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr4_+_175204865 | 0.14 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr15_+_41057818 | 0.14 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr13_-_30160925 | 0.14 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr1_+_79115503 | 0.14 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr9_-_5339873 | 0.13 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr2_+_183982255 | 0.13 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr4_+_66536248 | 0.13 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr9_+_130911770 | 0.13 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr17_-_57229155 | 0.12 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_136070614 | 0.12 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_+_209602156 | 0.12 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr12_-_50643664 | 0.12 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr9_+_130911723 | 0.12 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr20_+_43990576 | 0.11 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr19_-_17366257 | 0.11 |
ENST00000594059.1
|
AC010646.3
|
Uncharacterized protein |
| chr17_+_62223320 | 0.11 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr14_-_100046444 | 0.11 |
ENST00000554996.1
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr3_+_40518599 | 0.10 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr7_-_100860851 | 0.10 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr4_+_169575875 | 0.10 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_7118755 | 0.10 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr4_-_111563279 | 0.09 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr7_-_100239132 | 0.09 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr15_-_55562479 | 0.09 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_7895074 | 0.09 |
ENST00000270530.4
|
EVI5L
|
ecotropic viral integration site 5-like |
| chr7_-_99716952 | 0.09 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr10_+_99627889 | 0.09 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr17_-_27418537 | 0.09 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr1_-_68698197 | 0.08 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr21_+_47706537 | 0.08 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr1_-_201140673 | 0.08 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr3_-_121740969 | 0.08 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr15_+_84841242 | 0.08 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr10_-_22292675 | 0.08 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr4_+_144354644 | 0.08 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr7_-_99716940 | 0.08 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrX_+_43515467 | 0.08 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr7_+_99647389 | 0.08 |
ENST00000543588.1
ENST00000292450.4 ENST00000456748.2 |
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chr21_+_43823983 | 0.08 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr7_-_99717463 | 0.08 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr16_-_3350614 | 0.08 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr9_+_135937365 | 0.07 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr16_+_28763108 | 0.07 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr7_+_5919458 | 0.07 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chr12_-_123201337 | 0.07 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr4_-_176733897 | 0.07 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr9_+_124329336 | 0.07 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr3_-_100558953 | 0.07 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr6_-_62996066 | 0.07 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr8_+_22132810 | 0.07 |
ENST00000356766.6
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr4_+_123161120 | 0.07 |
ENST00000446180.1
|
KIAA1109
|
KIAA1109 |
| chr3_-_160823040 | 0.07 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr4_-_40632844 | 0.06 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr11_-_107729287 | 0.06 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_+_100860949 | 0.06 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr1_-_68698222 | 0.06 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chrX_-_13835147 | 0.06 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr20_-_44600810 | 0.06 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr1_-_52870104 | 0.06 |
ENST00000371568.3
|
ORC1
|
origin recognition complex, subunit 1 |
| chr7_-_99764907 | 0.06 |
ENST00000413800.1
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
| chr2_+_191208656 | 0.06 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr6_+_90192974 | 0.06 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr11_+_118398178 | 0.06 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr4_-_19458597 | 0.06 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr12_-_6484376 | 0.06 |
ENST00000360168.3
ENST00000358945.3 |
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr2_+_208423840 | 0.06 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_+_59279851 | 0.06 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr11_+_15095108 | 0.06 |
ENST00000324229.6
ENST00000533448.1 |
CALCB
|
calcitonin-related polypeptide beta |
| chr1_-_54879140 | 0.06 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr22_-_32651326 | 0.06 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr2_+_197577841 | 0.05 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr17_-_73258425 | 0.05 |
ENST00000578348.1
ENST00000582486.1 ENST00000582717.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr11_-_107729887 | 0.05 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr19_+_11071546 | 0.05 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr3_-_130745571 | 0.05 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr8_+_22132847 | 0.05 |
ENST00000521356.1
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr10_-_22292613 | 0.05 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr2_+_191208601 | 0.05 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr21_-_27423339 | 0.05 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr22_-_30960876 | 0.05 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_-_8113542 | 0.05 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr17_-_73901494 | 0.05 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr12_+_94071341 | 0.05 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chrX_-_24690771 | 0.05 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr19_+_50270219 | 0.05 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr11_-_107729504 | 0.05 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr22_-_29107919 | 0.05 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr7_+_99647411 | 0.05 |
ENST00000438937.1
|
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chr18_+_55888767 | 0.05 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_-_6737576 | 0.05 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chr16_-_21868739 | 0.05 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr16_-_21431078 | 0.05 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr14_+_61654271 | 0.05 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr12_+_56522001 | 0.05 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr12_+_128399965 | 0.05 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr9_+_116172958 | 0.05 |
ENST00000374165.1
|
C9orf43
|
chromosome 9 open reading frame 43 |
| chr16_-_31085514 | 0.05 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr14_-_24711806 | 0.05 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr16_-_28621312 | 0.05 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr16_+_50730910 | 0.05 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr19_+_54641444 | 0.05 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr16_-_21868978 | 0.05 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr4_+_119199904 | 0.05 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr2_+_234580499 | 0.05 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr17_-_62208169 | 0.05 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr2_-_113810444 | 0.04 |
ENST00000259213.4
ENST00000327407.2 |
IL36B
|
interleukin 36, beta |
| chr7_+_55980331 | 0.04 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr6_+_149721495 | 0.04 |
ENST00000326669.4
|
SUMO4
|
small ubiquitin-like modifier 4 |
| chr6_+_127898312 | 0.04 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_+_159750776 | 0.04 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr4_+_144312659 | 0.04 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr5_+_55149150 | 0.04 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr21_-_37914898 | 0.04 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr2_-_73869508 | 0.04 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr1_-_109935819 | 0.04 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr5_-_96478466 | 0.04 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr22_-_32766972 | 0.04 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr12_+_15699286 | 0.04 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr16_+_103816 | 0.04 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr1_+_156030937 | 0.04 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chrX_+_10031499 | 0.04 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr19_+_10362577 | 0.04 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr6_+_150690028 | 0.04 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr4_-_109541539 | 0.04 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr3_-_160823158 | 0.04 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr11_-_63376013 | 0.04 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr15_-_44969022 | 0.04 |
ENST00000560110.1
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr1_+_186265399 | 0.04 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr14_-_24711865 | 0.04 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_+_150639360 | 0.04 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr3_+_195447738 | 0.04 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr1_+_160370344 | 0.04 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr12_-_117175819 | 0.04 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr11_-_128894053 | 0.04 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr2_+_234580525 | 0.04 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr16_-_28374829 | 0.04 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr1_-_146040968 | 0.04 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr11_-_102714534 | 0.04 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr22_+_21996549 | 0.04 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr19_-_55677920 | 0.04 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr8_+_39972170 | 0.04 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr2_+_200820494 | 0.04 |
ENST00000435773.2
|
C2orf47
|
chromosome 2 open reading frame 47 |
| chr4_+_87515454 | 0.04 |
ENST00000427191.2
ENST00000436978.1 ENST00000502971.1 |
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr11_-_102709441 | 0.04 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr15_+_92006567 | 0.04 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr3_+_186288454 | 0.04 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr5_+_129083772 | 0.03 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr4_-_69111401 | 0.03 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr1_-_21620877 | 0.03 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr5_+_61874696 | 0.03 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr8_+_27168988 | 0.03 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_-_51796226 | 0.03 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr2_-_70780572 | 0.03 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr12_+_128399917 | 0.03 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr19_-_3557570 | 0.03 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr12_+_22778116 | 0.03 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr7_+_129015671 | 0.03 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_+_99202003 | 0.03 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr20_+_43991668 | 0.03 |
ENST00000243918.5
ENST00000426004.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr5_+_61874562 | 0.03 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr12_+_56521840 | 0.03 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr22_+_24199113 | 0.03 |
ENST00000405847.1
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr2_+_234545148 | 0.03 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr6_+_130339710 | 0.03 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_-_100565249 | 0.03 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr18_-_21891460 | 0.03 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr12_-_11339543 | 0.03 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr4_+_40198527 | 0.03 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr19_-_6424783 | 0.03 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr15_-_55562582 | 0.03 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_28618532 | 0.03 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr16_-_29415350 | 0.03 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr3_-_164875850 | 0.03 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr8_-_41166953 | 0.03 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr6_+_134274354 | 0.03 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr15_-_55563072 | 0.03 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_+_43152191 | 0.03 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr19_-_3985455 | 0.03 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr7_+_37723420 | 0.03 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr12_+_112563335 | 0.03 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr1_+_35258592 | 0.03 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr11_-_104905840 | 0.03 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr6_-_42690312 | 0.03 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr6_+_34204642 | 0.03 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |