Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
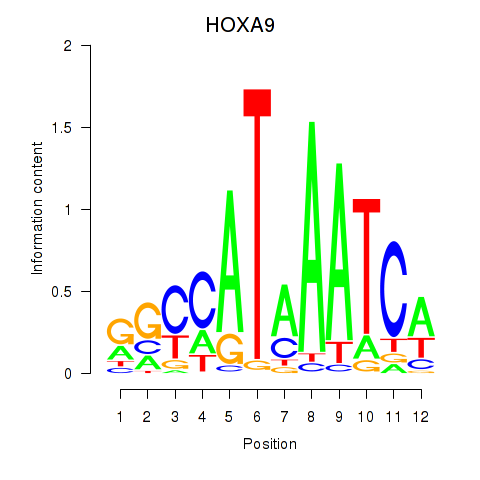
Results for HOXA9
Z-value: 0.59
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | homeobox A9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | 0.11 | 8.3e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_99578776 | 0.46 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr5_+_68485433 | 0.42 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr12_-_121476959 | 0.42 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr6_-_18249971 | 0.40 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr1_+_95616933 | 0.39 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr12_-_121477039 | 0.34 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr7_+_120629653 | 0.34 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_35013217 | 0.31 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr6_+_26104104 | 0.31 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr6_+_24126350 | 0.29 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr2_+_143635222 | 0.29 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr1_-_193075180 | 0.29 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr15_+_63414760 | 0.29 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr2_+_161993465 | 0.29 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_45629661 | 0.29 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr2_+_143635067 | 0.29 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr4_+_86748898 | 0.27 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_76439095 | 0.26 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr1_+_150254936 | 0.25 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr2_+_38177575 | 0.25 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr14_+_97925151 | 0.25 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chrX_-_119693745 | 0.24 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr14_-_81425828 | 0.24 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chrX_+_95939638 | 0.24 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr12_-_118796971 | 0.23 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr7_-_83824449 | 0.23 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_+_40028620 | 0.22 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr4_-_69536346 | 0.21 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr10_+_91092241 | 0.21 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr19_+_46367518 | 0.21 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr8_+_16884740 | 0.21 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr8_-_42358742 | 0.21 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr3_+_136649311 | 0.20 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr13_+_52586517 | 0.20 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr16_+_56691911 | 0.19 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chrX_+_95939711 | 0.19 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr3_-_8811288 | 0.18 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr19_-_6375860 | 0.18 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr16_+_56691838 | 0.18 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr12_-_13248598 | 0.17 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr1_-_220608023 | 0.17 |
ENST00000416839.2
|
AC096644.1
|
Uncharacterized protein |
| chr4_-_76439483 | 0.17 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr5_-_43515231 | 0.17 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr9_-_88356789 | 0.16 |
ENST00000357081.3
ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr11_-_10879572 | 0.16 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr18_-_68004529 | 0.16 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr12_+_59989918 | 0.15 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_99870744 | 0.15 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr5_-_111754948 | 0.15 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr16_+_56691606 | 0.15 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr1_-_197169672 | 0.15 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr18_+_9102669 | 0.15 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr10_+_15001430 | 0.15 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr1_+_61869748 | 0.15 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr3_+_87276407 | 0.15 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr2_+_161993412 | 0.15 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_108994466 | 0.15 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr19_-_52598958 | 0.15 |
ENST00000594440.1
ENST00000426391.2 ENST00000389534.4 |
ZNF841
|
zinc finger protein 841 |
| chr11_-_10879593 | 0.14 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr1_+_149230680 | 0.14 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chrX_-_63615297 | 0.14 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr18_+_1099004 | 0.14 |
ENST00000581556.1
|
RP11-78F17.1
|
RP11-78F17.1 |
| chr6_-_135818368 | 0.14 |
ENST00000367798.2
|
AHI1
|
Abelson helper integration site 1 |
| chr4_-_99578789 | 0.14 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr20_-_48782639 | 0.14 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr8_+_76452097 | 0.14 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr14_+_78174414 | 0.14 |
ENST00000557342.1
ENST00000238688.5 ENST00000557623.1 ENST00000557431.1 ENST00000556831.1 ENST00000556375.1 ENST00000553981.1 |
SLIRP
|
SRA stem-loop interacting RNA binding protein |
| chrX_+_119737806 | 0.14 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr12_-_121476750 | 0.14 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr7_-_27169801 | 0.14 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr7_-_99332719 | 0.14 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr11_-_104905840 | 0.14 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr3_-_197686847 | 0.14 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr1_+_52082751 | 0.13 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr3_+_119298523 | 0.13 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr5_+_34915444 | 0.13 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr2_+_196440692 | 0.13 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr4_+_39046615 | 0.13 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr19_-_18314836 | 0.13 |
ENST00000464076.3
ENST00000222256.4 |
RAB3A
|
RAB3A, member RAS oncogene family |
| chr2_+_182850551 | 0.13 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr15_-_41166414 | 0.13 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr6_+_47666275 | 0.13 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr5_+_159436120 | 0.13 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr17_-_8021710 | 0.12 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr8_+_31497271 | 0.12 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr6_-_131299929 | 0.12 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr11_-_10920714 | 0.12 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr11_-_14521379 | 0.12 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr14_+_101361107 | 0.12 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr12_-_88535747 | 0.12 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr15_+_93749295 | 0.12 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr1_+_198189921 | 0.12 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr6_+_99282570 | 0.12 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr6_+_75994755 | 0.12 |
ENST00000607799.1
|
RP1-234P15.4
|
RP1-234P15.4 |
| chr11_+_5646213 | 0.12 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr17_+_39421591 | 0.11 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr1_+_38512799 | 0.11 |
ENST00000432922.1
ENST00000428151.1 |
RP5-884C9.2
|
RP5-884C9.2 |
| chr21_-_35014027 | 0.11 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr8_+_52730143 | 0.11 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr13_+_27825446 | 0.11 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21 |
| chr12_+_122880045 | 0.11 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr3_-_52864680 | 0.11 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr3_+_108321623 | 0.11 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr1_-_95285652 | 0.11 |
ENST00000442418.1
|
LINC01057
|
long intergenic non-protein coding RNA 1057 |
| chr12_+_14956506 | 0.11 |
ENST00000330828.2
|
C12orf60
|
chromosome 12 open reading frame 60 |
| chr2_+_171571827 | 0.11 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr11_+_1430983 | 0.11 |
ENST00000524702.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr4_+_86749045 | 0.10 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_-_91618285 | 0.10 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr15_+_79165151 | 0.10 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr2_+_67624430 | 0.10 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr5_+_38845960 | 0.10 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr15_+_27216297 | 0.10 |
ENST00000333743.6
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
| chr11_-_8959758 | 0.10 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr6_-_137539651 | 0.10 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr3_+_157154578 | 0.10 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_2162162 | 0.10 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr1_-_85100703 | 0.10 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr4_-_74964904 | 0.10 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr5_+_177433406 | 0.10 |
ENST00000504530.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr5_-_148758839 | 0.10 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr15_-_54025300 | 0.10 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr8_-_97247759 | 0.09 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr22_+_22723969 | 0.09 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr2_+_10223908 | 0.09 |
ENST00000425235.1
|
AC104794.4
|
AC104794.4 |
| chr2_-_154335300 | 0.09 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr14_+_58894706 | 0.09 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr2_-_204400013 | 0.09 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr12_+_16500037 | 0.09 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_-_94301107 | 0.09 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr15_+_79165112 | 0.09 |
ENST00000426013.2
|
MORF4L1
|
mortality factor 4 like 1 |
| chr1_-_144866711 | 0.09 |
ENST00000530130.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_+_64069016 | 0.09 |
ENST00000488245.2
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_-_204399976 | 0.09 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr13_+_76445187 | 0.09 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr4_-_71532339 | 0.09 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr2_+_13677795 | 0.08 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr2_+_64069459 | 0.08 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr10_+_92631709 | 0.08 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr16_-_67427389 | 0.08 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr11_+_19138670 | 0.08 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr2_-_225434538 | 0.08 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr4_-_71532668 | 0.08 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chrX_+_114827818 | 0.08 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr17_+_58018269 | 0.08 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr1_-_33168336 | 0.08 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr7_-_83824169 | 0.08 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_70080449 | 0.08 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr11_+_57310114 | 0.08 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr14_+_45605127 | 0.08 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr14_+_61201445 | 0.08 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr12_-_54778444 | 0.07 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr4_-_48018680 | 0.07 |
ENST00000513178.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr9_-_127269661 | 0.07 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr2_-_166810261 | 0.07 |
ENST00000243344.7
|
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr7_-_75241096 | 0.07 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr1_+_104615595 | 0.07 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr4_+_71384300 | 0.07 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr1_-_152552980 | 0.07 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr1_+_44584522 | 0.07 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr2_+_233390890 | 0.07 |
ENST00000258385.3
ENST00000536614.1 ENST00000457943.2 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr7_+_65338312 | 0.07 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr11_-_133822111 | 0.07 |
ENST00000526663.1
|
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr11_+_34664014 | 0.07 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr3_-_149510553 | 0.07 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr5_-_150138061 | 0.07 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr10_-_71176623 | 0.07 |
ENST00000373306.4
|
TACR2
|
tachykinin receptor 2 |
| chrX_+_109602039 | 0.07 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr12_+_54402790 | 0.07 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr10_-_134599556 | 0.07 |
ENST00000368592.5
|
NKX6-2
|
NK6 homeobox 2 |
| chr7_+_93690435 | 0.07 |
ENST00000438538.1
|
AC003092.1
|
AC003092.1 |
| chr6_+_143447322 | 0.07 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr15_+_33022885 | 0.07 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr5_-_131347501 | 0.07 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_+_23749945 | 0.07 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr5_-_131347583 | 0.07 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_+_190541153 | 0.07 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr19_-_19739321 | 0.07 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr10_-_128210005 | 0.06 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr7_+_86781847 | 0.06 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr7_-_3214287 | 0.06 |
ENST00000404626.3
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr16_+_70613770 | 0.06 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr18_+_9102592 | 0.06 |
ENST00000318388.6
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr1_+_153940741 | 0.06 |
ENST00000431292.1
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr17_+_7758374 | 0.06 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr8_-_28347737 | 0.06 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr7_+_23749767 | 0.06 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr5_-_37371163 | 0.06 |
ENST00000513532.1
|
NUP155
|
nucleoporin 155kDa |
| chr3_+_142842128 | 0.06 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr5_+_75904950 | 0.06 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr16_+_19078960 | 0.06 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr5_-_37371278 | 0.06 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr5_+_140602904 | 0.06 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr1_-_89591749 | 0.06 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr12_-_14967095 | 0.06 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr16_+_19078911 | 0.06 |
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr10_+_74870206 | 0.06 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr10_+_79793518 | 0.06 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr11_+_85955787 | 0.05 |
ENST00000528180.1
|
EED
|
embryonic ectoderm development |
| chr18_-_10701979 | 0.05 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0039023 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:1990709 | maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0061571 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |