Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
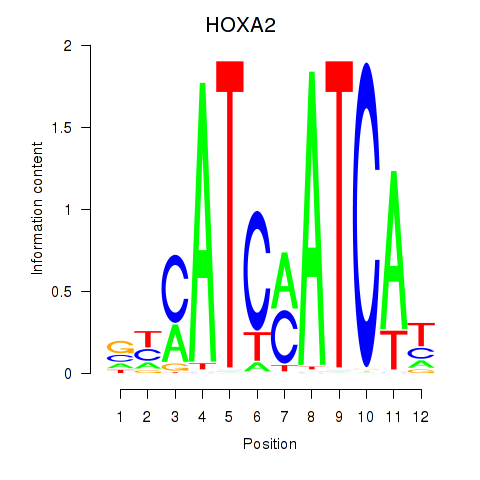
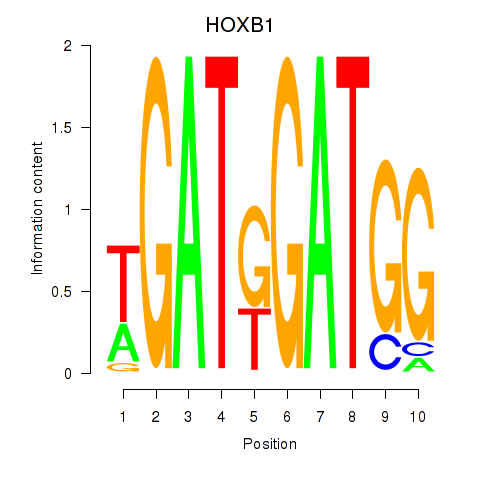
Results for HOXA2_HOXB1
Z-value: 0.26
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | 0.23 | 6.6e-01 | Click! |
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | 0.22 | 6.8e-01 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_162864575 | 0.27 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr17_+_7758374 | 0.20 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chrX_-_45629661 | 0.16 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr16_+_85936295 | 0.14 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr16_-_67427389 | 0.12 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr12_+_21679220 | 0.12 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr1_-_108507631 | 0.12 |
ENST00000527011.1
ENST00000370056.4 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr2_-_154335300 | 0.12 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr8_+_77596014 | 0.09 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr1_-_217804377 | 0.09 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr2_+_176995011 | 0.08 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chrX_+_95939638 | 0.08 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr11_+_133902547 | 0.08 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr7_-_27170352 | 0.08 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr5_+_98109322 | 0.08 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chrX_+_95939711 | 0.08 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr16_-_49890016 | 0.07 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr1_-_65533390 | 0.07 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr17_-_46623441 | 0.07 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr8_-_79717750 | 0.07 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr7_-_14029283 | 0.07 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr2_+_121493717 | 0.06 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr9_+_40028620 | 0.06 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr10_-_46342921 | 0.06 |
ENST00000448048.2
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr3_+_159481988 | 0.06 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr10_+_89264625 | 0.06 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr3_-_98619999 | 0.05 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr12_+_57390745 | 0.05 |
ENST00000600202.1
|
HBCBP
|
HBcAg-binding protein; Uncharacterized protein |
| chr11_-_2162162 | 0.05 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr7_+_79765071 | 0.05 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr15_-_93616340 | 0.05 |
ENST00000557420.1
ENST00000542321.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr7_-_14029515 | 0.05 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr9_-_101471479 | 0.04 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr19_-_45735138 | 0.04 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr11_-_126174186 | 0.04 |
ENST00000524964.1
|
RP11-712L6.5
|
Uncharacterized protein |
| chr12_-_28125638 | 0.04 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr15_+_75335604 | 0.04 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr8_-_67874805 | 0.04 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chrX_+_99899180 | 0.04 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr3_+_112929850 | 0.04 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_+_57482877 | 0.04 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr17_+_43239191 | 0.04 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr7_-_120497178 | 0.03 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr17_-_37353950 | 0.03 |
ENST00000394310.3
ENST00000394303.3 ENST00000344140.5 |
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr1_-_109584716 | 0.03 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr1_+_52082751 | 0.03 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr1_+_67390578 | 0.03 |
ENST00000371018.3
ENST00000355977.6 ENST00000357692.2 ENST00000401041.1 ENST00000371016.1 ENST00000371014.1 ENST00000371012.2 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr6_+_32006159 | 0.03 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr18_+_18943554 | 0.03 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr10_-_65028817 | 0.03 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr8_+_22250334 | 0.03 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr1_-_43833628 | 0.03 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr15_-_74501360 | 0.03 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr5_+_167181917 | 0.03 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr8_+_143530791 | 0.03 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr6_+_73331520 | 0.03 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr16_+_66429358 | 0.02 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr19_+_44645731 | 0.02 |
ENST00000426739.2
|
ZNF234
|
zinc finger protein 234 |
| chr2_-_242089677 | 0.02 |
ENST00000405260.1
|
PASK
|
PAS domain containing serine/threonine kinase |
| chr6_-_64029879 | 0.02 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr12_-_28124903 | 0.02 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr10_-_98945515 | 0.02 |
ENST00000371070.4
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr17_+_43239231 | 0.02 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr10_-_65028938 | 0.02 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr10_+_24755416 | 0.02 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr10_-_79397547 | 0.02 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_-_159065741 | 0.02 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr19_+_52074502 | 0.02 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr13_+_36050881 | 0.02 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr19_+_37997812 | 0.02 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr2_-_145277882 | 0.02 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_189507432 | 0.02 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr9_-_26947453 | 0.02 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr11_-_111782484 | 0.02 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr2_+_12857015 | 0.02 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr2_+_114647504 | 0.02 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr7_-_78400598 | 0.02 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_-_98945264 | 0.02 |
ENST00000314867.5
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr16_-_11617444 | 0.02 |
ENST00000598234.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr5_+_145826867 | 0.02 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr7_-_100881109 | 0.02 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr18_-_53070913 | 0.01 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr2_-_200323414 | 0.01 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr9_+_113431029 | 0.01 |
ENST00000189978.5
ENST00000374448.4 ENST00000374440.3 |
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr12_-_57472522 | 0.01 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr7_-_27142290 | 0.01 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr2_+_242089833 | 0.01 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr3_+_189507523 | 0.01 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr11_-_93583697 | 0.01 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr2_+_26624775 | 0.01 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr6_-_113754604 | 0.01 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr1_+_178062855 | 0.01 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_44588694 | 0.01 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr19_+_44645700 | 0.01 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr7_+_1272522 | 0.01 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr14_-_24733444 | 0.01 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr10_-_79397316 | 0.01 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr19_-_49944806 | 0.01 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr15_+_81591757 | 0.01 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr22_-_39151463 | 0.01 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr11_-_111782696 | 0.01 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr12_+_56414795 | 0.01 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr2_-_44588679 | 0.01 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr3_+_239652 | 0.01 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr3_-_72150076 | 0.01 |
ENST00000488545.1
ENST00000608654.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr9_-_127269661 | 0.01 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chrX_-_110655306 | 0.01 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr11_-_126870683 | 0.01 |
ENST00000525704.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr9_-_36276966 | 0.01 |
ENST00000543356.2
ENST00000396594.3 |
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr12_+_56414851 | 0.01 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr8_-_79717163 | 0.01 |
ENST00000520269.1
|
IL7
|
interleukin 7 |
| chr10_-_79397479 | 0.00 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_109584608 | 0.00 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr2_+_54684327 | 0.00 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr12_+_10658489 | 0.00 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr7_-_78400364 | 0.00 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_-_79397202 | 0.00 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_+_39975455 | 0.00 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr2_-_197226875 | 0.00 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_-_93616892 | 0.00 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr9_+_113431059 | 0.00 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr10_-_98945677 | 0.00 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr5_-_179045199 | 0.00 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr7_+_86273218 | 0.00 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr17_-_46608272 | 0.00 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr1_-_67390474 | 0.00 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr13_+_97928395 | 0.00 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr12_-_118490217 | 0.00 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr2_+_12857043 | 0.00 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chrX_-_110655391 | 0.00 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr17_+_39975544 | 0.00 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr12_+_7282795 | 0.00 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr4_+_129732467 | 0.00 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chrX_-_10645773 | 0.00 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |