Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
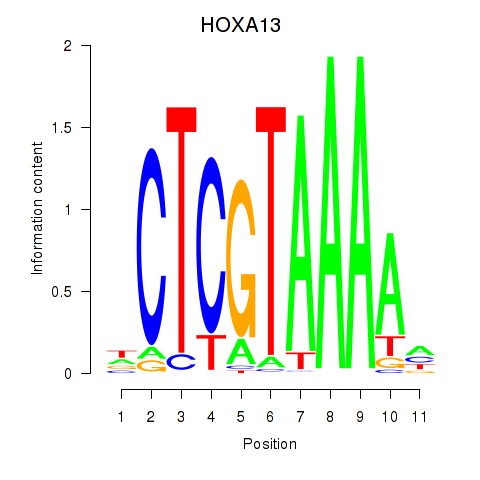
Results for HOXA13
Z-value: 0.64
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | homeobox A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA13 | hg19_v2_chr7_-_27239703_27239725 | 0.26 | 6.2e-01 | Click! |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_615942 | 0.73 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr14_-_88282595 | 0.42 |
ENST00000554519.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr7_-_92777606 | 0.34 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr5_-_78808617 | 0.34 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr10_-_61513146 | 0.31 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr12_-_116714564 | 0.25 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr19_-_7167989 | 0.23 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr8_+_77318769 | 0.23 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr13_+_78315528 | 0.22 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr15_+_85523671 | 0.21 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr22_+_18632666 | 0.21 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr15_-_79576156 | 0.18 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr11_+_29181503 | 0.18 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr19_-_44809121 | 0.17 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr8_+_35649365 | 0.17 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr22_-_29107919 | 0.17 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr4_+_54243917 | 0.17 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr16_+_86612112 | 0.16 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr1_-_156269428 | 0.15 |
ENST00000339922.3
|
VHLL
|
von Hippel-Lindau tumor suppressor-like |
| chr7_+_23719749 | 0.15 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr2_+_33359473 | 0.15 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr16_-_90096309 | 0.14 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr3_+_113251143 | 0.14 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr7_-_41742697 | 0.14 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr14_-_71067360 | 0.14 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr4_-_155533787 | 0.14 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr5_-_39270725 | 0.14 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr7_-_17598506 | 0.13 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr15_+_79166065 | 0.13 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr22_+_20193905 | 0.13 |
ENST00000609602.1
|
LINC00896
|
long intergenic non-protein coding RNA 896 |
| chr6_+_30875955 | 0.13 |
ENST00000259895.4
ENST00000539324.1 ENST00000376316.2 ENST00000453897.2 |
GTF2H4
|
general transcription factor IIH, polypeptide 4, 52kDa |
| chr13_+_78315466 | 0.12 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_-_123339343 | 0.12 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr8_+_94710789 | 0.12 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr4_+_54243798 | 0.12 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr2_+_162087577 | 0.12 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_-_124081008 | 0.12 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr6_+_134273321 | 0.12 |
ENST00000457715.1
|
TBPL1
|
TBP-like 1 |
| chr16_+_75252878 | 0.11 |
ENST00000361017.4
|
CTRB1
|
chymotrypsinogen B1 |
| chr20_-_50722183 | 0.11 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr20_-_44420507 | 0.11 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr3_+_111578027 | 0.11 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr17_-_39526052 | 0.11 |
ENST00000251646.3
|
KRT33B
|
keratin 33B |
| chr10_-_61513201 | 0.11 |
ENST00000414264.1
ENST00000594536.1 |
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr6_+_121756809 | 0.11 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr3_+_57882061 | 0.11 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr12_+_75874580 | 0.11 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr4_+_54243862 | 0.11 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr11_-_61849656 | 0.11 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr4_-_83280767 | 0.10 |
ENST00000514671.1
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr3_-_118753792 | 0.10 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr2_+_33661382 | 0.10 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr16_-_46865286 | 0.10 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr11_+_101983176 | 0.10 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr14_+_24701819 | 0.10 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr5_+_66254698 | 0.10 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_192109911 | 0.10 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr2_-_122262593 | 0.10 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr3_+_50211240 | 0.10 |
ENST00000420831.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr10_+_51371390 | 0.10 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr14_+_61201445 | 0.10 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr17_-_39538550 | 0.10 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr16_-_28192360 | 0.09 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr17_-_40169659 | 0.09 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr17_-_46688334 | 0.09 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr6_+_106988986 | 0.09 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr7_+_129015671 | 0.09 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_-_31845914 | 0.09 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr14_+_56078695 | 0.09 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr2_+_33359687 | 0.09 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr14_-_92414055 | 0.09 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr3_+_101546827 | 0.08 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr7_+_23749945 | 0.08 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr12_+_28605426 | 0.08 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr4_+_122722466 | 0.08 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr9_-_136039282 | 0.08 |
ENST00000372036.3
ENST00000372038.3 ENST00000540636.1 ENST00000372043.3 ENST00000542690.1 |
GBGT1
RALGDS
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 ral guanine nucleotide dissociation stimulator |
| chr13_+_78315348 | 0.08 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_+_207804278 | 0.08 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr11_+_101785727 | 0.08 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr6_+_4706368 | 0.08 |
ENST00000328908.5
|
CDYL
|
chromodomain protein, Y-like |
| chr5_-_150284351 | 0.08 |
ENST00000427179.1
|
ZNF300
|
zinc finger protein 300 |
| chr7_-_151511911 | 0.08 |
ENST00000392801.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr14_+_24702099 | 0.08 |
ENST00000420554.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr7_-_69062391 | 0.07 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr1_-_150208320 | 0.07 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr2_+_33359646 | 0.07 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr20_+_55204351 | 0.07 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr15_+_52155001 | 0.07 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr3_+_57875738 | 0.07 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr12_-_56367101 | 0.07 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr14_+_24701870 | 0.07 |
ENST00000561035.1
ENST00000559409.1 ENST00000558865.1 ENST00000558279.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr20_+_37554955 | 0.07 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr3_-_126373929 | 0.07 |
ENST00000523403.1
ENST00000524230.2 |
TXNRD3
|
thioredoxin reductase 3 |
| chr13_+_34392200 | 0.07 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr17_+_57233087 | 0.07 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr7_-_27224795 | 0.07 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr12_-_74796291 | 0.07 |
ENST00000551726.1
|
RP11-81H3.2
|
RP11-81H3.2 |
| chr9_+_137979506 | 0.07 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr10_+_103986085 | 0.07 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr12_+_12878829 | 0.07 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_-_150208363 | 0.07 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_68860949 | 0.07 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr17_-_73150629 | 0.07 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr16_+_53483983 | 0.07 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chrX_-_41449204 | 0.07 |
ENST00000378179.3
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr16_+_19619083 | 0.07 |
ENST00000538552.1
|
C16orf62
|
chromosome 16 open reading frame 62 |
| chr14_+_24701628 | 0.07 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_+_199996702 | 0.07 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_+_111765562 | 0.07 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr20_+_37590942 | 0.07 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr5_+_158737824 | 0.06 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr15_+_71228826 | 0.06 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_+_24702073 | 0.06 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr5_+_159895275 | 0.06 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr1_-_150208291 | 0.06 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_+_77030267 | 0.06 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_+_37571717 | 0.06 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr3_-_156878482 | 0.06 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr8_+_133879193 | 0.06 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr5_+_111964133 | 0.06 |
ENST00000508879.1
ENST00000507565.1 |
RP11-159K7.2
|
RP11-159K7.2 |
| chr9_-_2844058 | 0.06 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr14_+_24702127 | 0.06 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_+_19967014 | 0.06 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr9_+_112887772 | 0.05 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr5_+_138210919 | 0.05 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr22_+_39916558 | 0.05 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr3_-_112013071 | 0.05 |
ENST00000487372.1
ENST00000486574.1 ENST00000305815.5 |
SLC9C1
|
solute carrier family 9, subfamily C (Na+-transporting carboxylic acid decarboxylase), member 1 |
| chr5_+_95997885 | 0.05 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr16_+_28565230 | 0.05 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr10_+_106028923 | 0.05 |
ENST00000338595.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr2_-_169104651 | 0.05 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr2_+_64681103 | 0.05 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr14_-_92414294 | 0.05 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr3_+_57882024 | 0.05 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr7_+_23749894 | 0.05 |
ENST00000433467.2
|
STK31
|
serine/threonine kinase 31 |
| chr4_+_106631966 | 0.05 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr11_+_34999328 | 0.05 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr12_-_89413456 | 0.05 |
ENST00000500381.2
|
RP11-13A1.1
|
RP11-13A1.1 |
| chr8_-_110346614 | 0.05 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr17_-_56769382 | 0.05 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr10_-_51623203 | 0.05 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr5_-_138533973 | 0.05 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr5_-_74532696 | 0.05 |
ENST00000506364.2
ENST00000274361.3 |
ANKRD31
|
ankyrin repeat domain 31 |
| chr19_+_852291 | 0.05 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr12_-_50677255 | 0.05 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr15_-_41836441 | 0.05 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr18_-_53255766 | 0.05 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr3_+_158449972 | 0.05 |
ENST00000486568.1
|
MFSD1
|
major facilitator superfamily domain containing 1 |
| chrX_+_9431324 | 0.05 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr2_+_202047596 | 0.05 |
ENST00000286186.6
ENST00000360132.3 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr9_+_706842 | 0.05 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr2_+_189839046 | 0.04 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr19_+_13858701 | 0.04 |
ENST00000586666.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chrX_+_117480036 | 0.04 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr6_-_33041378 | 0.04 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr2_+_114195268 | 0.04 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr3_+_167582561 | 0.04 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr8_-_87242589 | 0.04 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr14_+_96722539 | 0.04 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr11_-_4719072 | 0.04 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr1_-_150208412 | 0.04 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr19_-_49496557 | 0.04 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr14_+_74353320 | 0.04 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr21_+_39628655 | 0.04 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr21_+_17792672 | 0.04 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_+_91933726 | 0.04 |
ENST00000534113.2
|
SECISBP2
|
SECIS binding protein 2 |
| chr5_-_179050660 | 0.04 |
ENST00000519056.1
ENST00000506721.1 ENST00000503105.1 ENST00000504348.1 ENST00000508103.1 ENST00000510431.1 ENST00000515158.1 ENST00000393432.4 ENST00000442819.2 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr12_-_52779433 | 0.04 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr4_-_120243545 | 0.04 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr19_+_36426452 | 0.04 |
ENST00000588831.1
|
LRFN3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr11_+_34460447 | 0.04 |
ENST00000241052.4
|
CAT
|
catalase |
| chr3_-_118753716 | 0.04 |
ENST00000393775.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr3_-_118753626 | 0.04 |
ENST00000489689.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr2_+_12858355 | 0.04 |
ENST00000405331.3
|
TRIB2
|
tribbles pseudokinase 2 |
| chr19_+_50084561 | 0.04 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr20_-_52687059 | 0.04 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr12_-_47473425 | 0.04 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr2_+_170366203 | 0.04 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr17_-_15496722 | 0.04 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr1_+_222886694 | 0.04 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr3_-_123680047 | 0.04 |
ENST00000409697.3
|
CCDC14
|
coiled-coil domain containing 14 |
| chr5_-_150284532 | 0.04 |
ENST00000394226.2
ENST00000446148.2 ENST00000274599.5 ENST00000418587.2 |
ZNF300
|
zinc finger protein 300 |
| chr2_+_32390925 | 0.03 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr16_+_68119764 | 0.03 |
ENST00000570212.1
ENST00000562926.1 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr13_+_34392185 | 0.03 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr17_-_40021656 | 0.03 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr14_-_50319758 | 0.03 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr7_+_23749767 | 0.03 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr18_+_60206744 | 0.03 |
ENST00000586834.1
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr7_+_23749854 | 0.03 |
ENST00000456014.2
|
STK31
|
serine/threonine kinase 31 |
| chr6_+_71377567 | 0.03 |
ENST00000422334.2
|
SMAP1
|
small ArfGAP 1 |
| chr17_-_7232585 | 0.03 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr3_-_99569821 | 0.03 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr14_+_23654525 | 0.03 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr5_-_58571935 | 0.03 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_-_150208498 | 0.03 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_125775629 | 0.03 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr8_+_26150628 | 0.03 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr5_+_95385819 | 0.03 |
ENST00000507997.1
|
RP11-254I22.1
|
RP11-254I22.1 |
| chr7_+_23749822 | 0.03 |
ENST00000422637.1
|
STK31
|
serine/threonine kinase 31 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:2000987 | cell communication by chemical coupling(GO:0010643) positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:1904585 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.0 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP in extracellular matrix(GO:0035582) sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |