Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
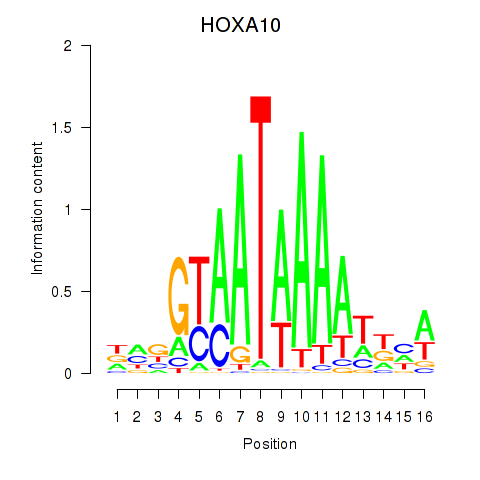
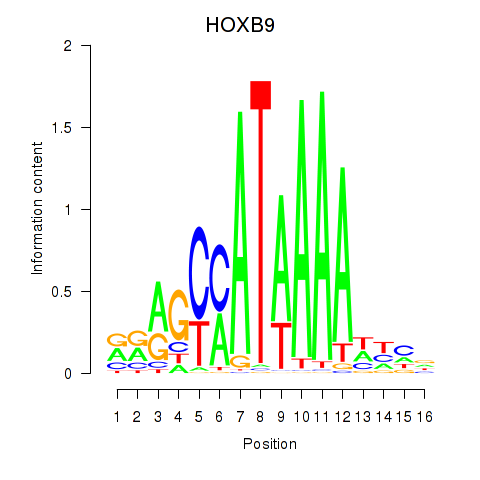
Results for HOXA10_HOXB9
Z-value: 0.50
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA10
|
ENSG00000253293.3 | homeobox A10 |
|
HOXB9
|
ENSG00000170689.8 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | 0.41 | 4.2e-01 | Click! |
| HOXA10 | hg19_v2_chr7_-_27219849_27219880 | -0.26 | 6.2e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_48683188 | 0.43 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr6_+_26156551 | 0.30 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr4_+_70894130 | 0.30 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr1_+_151735431 | 0.26 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr17_-_38821373 | 0.26 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr15_-_55541227 | 0.24 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_21106081 | 0.23 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr1_+_151739131 | 0.22 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr17_+_61151306 | 0.22 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_+_58166726 | 0.21 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr22_-_23922448 | 0.19 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr19_-_53636125 | 0.18 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr11_+_113185778 | 0.18 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr5_+_158737824 | 0.17 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr22_-_23922410 | 0.17 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr10_-_103599591 | 0.16 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr1_-_154178803 | 0.16 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr22_-_38240316 | 0.15 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr9_+_82188077 | 0.15 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_+_29704142 | 0.14 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr6_+_12958137 | 0.14 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr2_+_172309634 | 0.14 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr3_+_164924716 | 0.14 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr19_-_19626838 | 0.12 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr8_+_66955648 | 0.12 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr5_+_98109322 | 0.12 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr15_-_98417780 | 0.11 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr15_+_40886199 | 0.11 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr2_+_181988560 | 0.11 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr16_-_25122735 | 0.11 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr2_+_20101786 | 0.10 |
ENST00000607190.1
|
RP11-79O8.1
|
RP11-79O8.1 |
| chrX_+_105936982 | 0.10 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr2_+_38177575 | 0.10 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr17_-_29624343 | 0.10 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr4_-_71532207 | 0.10 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_84630574 | 0.10 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_101591314 | 0.09 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_+_208423840 | 0.09 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr7_-_81635106 | 0.09 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr10_-_73497581 | 0.09 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr1_-_160231451 | 0.08 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_-_161207986 | 0.08 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_+_71017791 | 0.08 |
ENST00000502294.1
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chrX_+_105937068 | 0.08 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr12_+_90313394 | 0.08 |
ENST00000549551.1
|
RP11-654D12.2
|
RP11-654D12.2 |
| chr15_+_40886439 | 0.08 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr1_+_43613612 | 0.08 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr16_+_25123148 | 0.07 |
ENST00000570981.1
|
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr14_+_73706308 | 0.07 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr9_+_34652164 | 0.07 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr1_-_226926864 | 0.07 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr20_+_42984330 | 0.07 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr9_-_104198042 | 0.07 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr1_+_12524965 | 0.06 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr9_+_67977438 | 0.06 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr11_-_104905840 | 0.06 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr9_+_131062367 | 0.06 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr9_+_130026756 | 0.06 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_+_107364769 | 0.06 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr9_-_133814455 | 0.06 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr9_+_134065506 | 0.06 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr10_-_99030395 | 0.06 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr12_+_75728419 | 0.06 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr9_-_133814527 | 0.06 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr12_-_21928515 | 0.06 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr10_+_135160844 | 0.06 |
ENST00000423766.1
ENST00000458230.1 |
PRAP1
|
proline-rich acidic protein 1 |
| chr8_+_145215928 | 0.06 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr10_+_69865866 | 0.06 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr4_+_70916119 | 0.06 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr16_+_66613351 | 0.06 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr15_+_75639372 | 0.05 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr19_-_9896672 | 0.05 |
ENST00000589412.1
ENST00000586814.1 |
ZNF846
|
zinc finger protein 846 |
| chr3_+_164924769 | 0.05 |
ENST00000494915.1
|
RP11-85M11.2
|
RP11-85M11.2 |
| chr22_+_43011247 | 0.05 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr14_-_64846033 | 0.05 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr5_-_87516448 | 0.05 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr19_-_19626351 | 0.05 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr14_-_38028689 | 0.05 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr19_-_3500635 | 0.05 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr1_+_67773044 | 0.05 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr3_-_172241250 | 0.04 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr4_+_71384300 | 0.04 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr17_-_46806540 | 0.04 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr8_-_8318847 | 0.04 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr6_+_25279759 | 0.04 |
ENST00000377969.3
|
LRRC16A
|
leucine rich repeat containing 16A |
| chr4_+_169575875 | 0.04 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_-_10115007 | 0.04 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr9_-_95640218 | 0.04 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr9_+_77112244 | 0.04 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr8_-_110660975 | 0.04 |
ENST00000528045.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr4_-_170897045 | 0.04 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr7_-_130080977 | 0.04 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr7_-_122840015 | 0.04 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr17_-_38928414 | 0.04 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr2_-_158182322 | 0.04 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr9_+_123884038 | 0.04 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr19_+_1440838 | 0.04 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr1_+_104615595 | 0.04 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr5_-_86534822 | 0.04 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr18_-_73967160 | 0.04 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr10_+_24755416 | 0.03 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr19_+_3185910 | 0.03 |
ENST00000588428.1
|
NCLN
|
nicalin |
| chr6_+_80816342 | 0.03 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr12_-_49581152 | 0.03 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr13_+_28813645 | 0.03 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr15_+_75639296 | 0.03 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_+_154401791 | 0.03 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr2_+_27440229 | 0.03 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr22_-_43010859 | 0.03 |
ENST00000339677.6
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr9_+_95736758 | 0.03 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr1_-_23520755 | 0.03 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr4_-_36245561 | 0.03 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_127710292 | 0.03 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr2_+_28718921 | 0.03 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr6_-_31830655 | 0.03 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr1_-_95846528 | 0.03 |
ENST00000427695.3
|
RP11-14O19.2
|
RP11-14O19.2 |
| chr8_+_77593448 | 0.03 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr5_+_140261703 | 0.03 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr16_-_48281444 | 0.03 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr11_-_104916034 | 0.03 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr9_-_85678043 | 0.03 |
ENST00000376447.3
ENST00000340717.4 |
RASEF
|
RAS and EF-hand domain containing |
| chr20_-_36152914 | 0.03 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr22_-_31324215 | 0.03 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr19_+_19626531 | 0.03 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr4_-_139051839 | 0.03 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr22_+_24407642 | 0.03 |
ENST00000454754.1
ENST00000263119.5 |
CABIN1
|
calcineurin binding protein 1 |
| chr7_-_143105941 | 0.03 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr10_-_73848086 | 0.03 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr6_+_83903061 | 0.03 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr8_+_91233711 | 0.03 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr4_-_104021009 | 0.02 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr8_+_21906658 | 0.02 |
ENST00000523300.1
|
DMTN
|
dematin actin binding protein |
| chr7_+_128784712 | 0.02 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr14_+_57671888 | 0.02 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chrX_-_117119243 | 0.02 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chrY_-_20935572 | 0.02 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr21_+_34398153 | 0.02 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr4_-_87281224 | 0.02 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr9_+_103947311 | 0.02 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr2_-_207078086 | 0.02 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr20_+_3767547 | 0.02 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr6_+_28249299 | 0.02 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr3_-_139396853 | 0.02 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_+_102927962 | 0.02 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr20_+_5987890 | 0.02 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_-_220263096 | 0.02 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr7_-_7782204 | 0.02 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr15_+_57891609 | 0.02 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr21_-_28215332 | 0.02 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr20_+_30697298 | 0.02 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr12_-_76879852 | 0.02 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr19_+_4402659 | 0.02 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chrX_+_73164149 | 0.02 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr7_+_57509877 | 0.02 |
ENST00000420713.1
|
ZNF716
|
zinc finger protein 716 |
| chr16_-_21663950 | 0.02 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr12_+_70219052 | 0.02 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr10_+_133747955 | 0.02 |
ENST00000455566.1
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr18_-_74839891 | 0.02 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr6_+_32812568 | 0.02 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_-_155880672 | 0.02 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr17_-_10560619 | 0.02 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr15_+_67418047 | 0.02 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr16_-_66907139 | 0.02 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr1_-_115301235 | 0.02 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr5_+_67586465 | 0.02 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr9_+_124413873 | 0.02 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr3_-_50375657 | 0.02 |
ENST00000395126.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr2_-_69180083 | 0.02 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr15_+_101417919 | 0.02 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chrY_+_20708557 | 0.02 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr2_-_122262593 | 0.02 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr9_-_20382446 | 0.02 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr1_+_73771844 | 0.02 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr9_-_104145795 | 0.02 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr19_-_29704448 | 0.02 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr12_+_116985896 | 0.02 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr22_-_32651326 | 0.02 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chrX_+_41548259 | 0.02 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr15_+_66797627 | 0.02 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr5_+_140165876 | 0.02 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr6_+_43737939 | 0.02 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chrX_-_135338503 | 0.02 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chrX_-_73072534 | 0.02 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr17_-_33469299 | 0.02 |
ENST00000586869.1
ENST00000360831.5 ENST00000442241.4 |
NLE1
|
notchless homolog 1 (Drosophila) |
| chr4_-_69536346 | 0.02 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr3_+_89156799 | 0.02 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr11_+_61717279 | 0.02 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chrX_-_54070388 | 0.02 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr3_+_125687987 | 0.02 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr5_-_20575959 | 0.02 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr2_+_220143989 | 0.02 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr4_-_140222358 | 0.02 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr6_+_28249332 | 0.02 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr20_-_25320367 | 0.02 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr8_+_146024261 | 0.01 |
ENST00000359971.3
ENST00000528012.1 |
ZNF517
|
zinc finger protein 517 |
| chr8_+_1993152 | 0.01 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr2_-_69180012 | 0.01 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr12_-_12491608 | 0.01 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr3_+_187957646 | 0.01 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.0 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |