Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
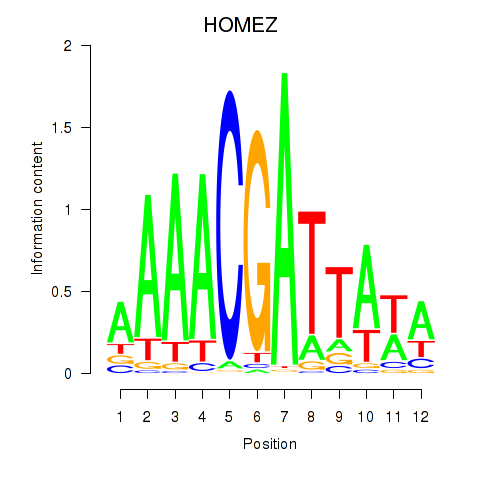
Results for HOMEZ
Z-value: 0.91
Transcription factors associated with HOMEZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOMEZ
|
ENSG00000215271.6 | homeobox and leucine zipper encoding |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOMEZ | hg19_v2_chr14_-_23762777_23762821 | 0.25 | 6.4e-01 | Click! |
Activity profile of HOMEZ motif
Sorted Z-values of HOMEZ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_116595028 | 0.99 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr19_+_44617511 | 0.63 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr17_+_73539232 | 0.56 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr8_-_65730127 | 0.55 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr19_-_52643157 | 0.54 |
ENST00000597013.1
ENST00000600228.1 ENST00000596290.1 |
ZNF616
|
zinc finger protein 616 |
| chr17_-_61045902 | 0.52 |
ENST00000581596.1
|
RP11-180P8.3
|
RP11-180P8.3 |
| chr12_+_11187087 | 0.47 |
ENST00000601123.1
|
AC018630.1
|
Uncharacterized protein |
| chr14_+_39944025 | 0.44 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr3_-_196242233 | 0.43 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr6_+_116601330 | 0.41 |
ENST00000449314.1
ENST00000453463.1 |
RP1-93H18.1
|
RP1-93H18.1 |
| chr6_-_13290684 | 0.40 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr6_+_111408698 | 0.37 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr6_-_27880174 | 0.37 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr18_+_44526786 | 0.37 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr6_+_31105426 | 0.36 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr6_-_137539651 | 0.36 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr4_-_112993808 | 0.35 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr11_+_115498761 | 0.34 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr6_-_137540477 | 0.33 |
ENST00000367735.2
ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1
|
interferon gamma receptor 1 |
| chrX_-_27417088 | 0.33 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr19_-_53400813 | 0.33 |
ENST00000595635.1
ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320
|
zinc finger protein 320 |
| chr12_+_9822331 | 0.31 |
ENST00000545918.1
ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chr12_+_54366894 | 0.30 |
ENST00000546378.1
ENST00000243082.4 |
HOXC11
|
homeobox C11 |
| chr1_+_158985457 | 0.30 |
ENST00000567661.1
ENST00000474473.1 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_81001398 | 0.30 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr18_+_44526744 | 0.29 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr12_-_42631529 | 0.29 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr3_-_107941209 | 0.29 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr6_+_27925019 | 0.29 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr12_-_112450915 | 0.29 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chr7_+_56032652 | 0.28 |
ENST00000437587.1
|
GBAS
|
glioblastoma amplified sequence |
| chr17_-_191188 | 0.28 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr19_-_36304201 | 0.28 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr16_-_75467318 | 0.27 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr15_+_63414760 | 0.27 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr1_+_104104379 | 0.27 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr10_+_13628933 | 0.27 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr9_-_95055923 | 0.26 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr22_-_28316116 | 0.25 |
ENST00000415296.1
|
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr7_-_48068643 | 0.25 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr1_-_62190793 | 0.25 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr7_-_102985035 | 0.25 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr2_+_210518057 | 0.25 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr16_+_33204156 | 0.24 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr2_-_234475380 | 0.24 |
ENST00000443711.2
ENST00000251722.6 |
USP40
|
ubiquitin specific peptidase 40 |
| chr11_-_8964580 | 0.24 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chrX_+_49178536 | 0.23 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr21_+_17909594 | 0.23 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_37068530 | 0.22 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr19_-_48389651 | 0.22 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr12_+_9822293 | 0.21 |
ENST00000261340.7
ENST00000290855.6 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chr1_+_44401479 | 0.21 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr9_-_100684769 | 0.20 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr3_-_146262637 | 0.20 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr18_-_46895066 | 0.20 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr3_-_146262352 | 0.20 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr9_-_32552551 | 0.19 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr8_+_86089460 | 0.19 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr6_+_79577189 | 0.19 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr10_-_126849626 | 0.19 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr10_+_114135952 | 0.18 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr2_-_190627481 | 0.18 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr4_+_47487285 | 0.18 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr2_-_112237835 | 0.18 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr20_+_44650348 | 0.18 |
ENST00000454036.2
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr12_+_19282643 | 0.18 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_16793160 | 0.18 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr1_+_63989004 | 0.18 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr8_-_123793048 | 0.17 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr3_-_124839648 | 0.17 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr4_-_164265984 | 0.17 |
ENST00000511901.1
|
NPY1R
|
neuropeptide Y receptor Y1 |
| chr12_+_133614062 | 0.17 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr9_+_104296163 | 0.17 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr2_+_191334212 | 0.17 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chrX_+_41583408 | 0.17 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chrX_+_86772707 | 0.17 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr2_-_14541060 | 0.17 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr17_-_57970074 | 0.16 |
ENST00000346141.6
|
TUBD1
|
tubulin, delta 1 |
| chr2_+_85843252 | 0.16 |
ENST00000409025.1
ENST00000409470.1 ENST00000323701.6 ENST00000409766.3 |
USP39
|
ubiquitin specific peptidase 39 |
| chr7_-_36406750 | 0.16 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr19_-_52408257 | 0.16 |
ENST00000354957.3
ENST00000600738.1 ENST00000595418.1 ENST00000599530.1 |
ZNF649
|
zinc finger protein 649 |
| chr7_+_89783689 | 0.16 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr11_+_14926543 | 0.16 |
ENST00000523376.1
|
CALCB
|
calcitonin-related polypeptide beta |
| chr15_+_69857515 | 0.16 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr6_-_27840099 | 0.15 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr10_+_95326416 | 0.15 |
ENST00000371481.4
ENST00000371483.4 ENST00000604414.1 |
FFAR4
|
free fatty acid receptor 4 |
| chr11_+_125464758 | 0.15 |
ENST00000529886.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr4_+_146560245 | 0.15 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr1_-_40349106 | 0.15 |
ENST00000545233.1
ENST00000537440.1 ENST00000537223.1 ENST00000541099.1 ENST00000441669.2 ENST00000544981.1 ENST00000316891.5 ENST00000372818.1 |
TRIT1
|
tRNA isopentenyltransferase 1 |
| chr15_-_74374891 | 0.15 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr6_+_107349392 | 0.15 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr19_+_21541777 | 0.15 |
ENST00000380870.4
ENST00000597810.1 ENST00000594245.1 |
ZNF738
|
zinc finger protein 738 |
| chr4_+_185395947 | 0.14 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr6_+_44355257 | 0.14 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr2_-_231860596 | 0.14 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr2_-_99917639 | 0.14 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr21_+_37692481 | 0.14 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr11_+_34664014 | 0.14 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr7_+_75024337 | 0.13 |
ENST00000450434.1
|
TRIM73
|
tripartite motif containing 73 |
| chr1_+_28099683 | 0.13 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr17_-_30677020 | 0.13 |
ENST00000583774.1
|
C17orf75
|
chromosome 17 open reading frame 75 |
| chr4_+_54243862 | 0.13 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr1_-_157789850 | 0.13 |
ENST00000491942.1
ENST00000358292.3 ENST00000368176.3 |
FCRL1
|
Fc receptor-like 1 |
| chr13_-_31038370 | 0.13 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr6_-_15548591 | 0.13 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr9_-_13279589 | 0.13 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr10_-_99052382 | 0.13 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr6_-_37225367 | 0.13 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr19_+_45418067 | 0.13 |
ENST00000589078.1
ENST00000586638.1 |
APOC1
|
apolipoprotein C-I |
| chr5_+_178450753 | 0.13 |
ENST00000444149.2
ENST00000519896.1 ENST00000522442.1 |
ZNF879
|
zinc finger protein 879 |
| chr15_-_90358564 | 0.12 |
ENST00000559874.1
|
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr3_+_108308559 | 0.12 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr7_+_99195677 | 0.12 |
ENST00000431679.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr21_+_40824003 | 0.12 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr2_+_183582774 | 0.12 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr19_-_9785724 | 0.12 |
ENST00000585350.1
ENST00000587392.1 ENST00000293648.4 |
ZNF562
|
zinc finger protein 562 |
| chr3_-_146262488 | 0.12 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr12_+_26126681 | 0.12 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chrX_+_1733876 | 0.12 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chrY_+_15016013 | 0.12 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr2_+_234590556 | 0.12 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr3_-_57678772 | 0.12 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr17_-_57970219 | 0.12 |
ENST00000394239.3
ENST00000539018.1 ENST00000340993.6 ENST00000325752.3 ENST00000590498.1 ENST00000592145.1 |
TUBD1
|
tubulin, delta 1 |
| chr12_-_64784306 | 0.11 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr2_+_61372226 | 0.11 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr6_+_116782527 | 0.11 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr8_-_101962777 | 0.11 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_-_107941230 | 0.11 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr8_+_28748765 | 0.11 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr3_+_133524459 | 0.11 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr3_-_155011483 | 0.11 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr4_-_90759440 | 0.11 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_-_54934708 | 0.11 |
ENST00000520534.1
ENST00000518784.1 ENST00000522635.1 |
TCEA1
|
transcription elongation factor A (SII), 1 |
| chr8_+_132916318 | 0.11 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_174670143 | 0.11 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr17_-_39183452 | 0.11 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr5_-_68664989 | 0.11 |
ENST00000508954.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr10_-_38265517 | 0.11 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr8_-_56986768 | 0.11 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chrX_+_86772787 | 0.11 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr17_-_36003487 | 0.11 |
ENST00000394367.3
ENST00000349699.2 |
DDX52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chr17_+_5389605 | 0.11 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr5_+_141488070 | 0.11 |
ENST00000253814.4
|
NDFIP1
|
Nedd4 family interacting protein 1 |
| chr19_-_46974741 | 0.11 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr4_+_95972822 | 0.11 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_44200146 | 0.10 |
ENST00000395510.2
ENST00000325127.4 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr19_+_44598534 | 0.10 |
ENST00000336976.6
|
ZNF224
|
zinc finger protein 224 |
| chr2_+_86947296 | 0.10 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr17_-_7167279 | 0.10 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr2_+_44502597 | 0.10 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr4_+_5712942 | 0.10 |
ENST00000509451.1
|
EVC
|
Ellis van Creveld syndrome |
| chr4_+_57276661 | 0.10 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr17_-_6554877 | 0.10 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chrX_-_135962876 | 0.10 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr14_-_91294472 | 0.10 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chrX_+_21959108 | 0.10 |
ENST00000457085.1
|
SMS
|
spermine synthase |
| chr1_+_20208870 | 0.10 |
ENST00000375120.3
|
OTUD3
|
OTU domain containing 3 |
| chr22_-_22901477 | 0.10 |
ENST00000420709.1
ENST00000398741.1 ENST00000405655.3 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr3_+_171561127 | 0.10 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr4_+_57845024 | 0.10 |
ENST00000431623.2
ENST00000441246.2 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr12_-_10978957 | 0.10 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr5_+_63461642 | 0.10 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr10_+_98064085 | 0.10 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr6_+_64281906 | 0.10 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_+_215740709 | 0.10 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr8_+_92082424 | 0.09 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chrX_+_70798261 | 0.09 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr1_+_28099700 | 0.09 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr6_-_37225391 | 0.09 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr15_+_72947079 | 0.09 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr17_+_5390220 | 0.09 |
ENST00000381165.3
|
MIS12
|
MIS12 kinetochore complex component |
| chr1_-_6259641 | 0.09 |
ENST00000234875.4
|
RPL22
|
ribosomal protein L22 |
| chr20_+_32951070 | 0.09 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr5_+_96038476 | 0.09 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr18_+_3247413 | 0.09 |
ENST00000579226.1
ENST00000217652.3 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_+_126102292 | 0.09 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr2_-_8977714 | 0.09 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr4_+_20702030 | 0.09 |
ENST00000510051.1
ENST00000503585.1 ENST00000360916.5 ENST00000295290.8 ENST00000514485.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr7_-_22539771 | 0.09 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr16_+_85832146 | 0.09 |
ENST00000565078.1
|
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr17_+_19091325 | 0.09 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr20_-_45280091 | 0.09 |
ENST00000396360.1
ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr9_-_13279563 | 0.09 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr3_-_57260377 | 0.09 |
ENST00000495160.2
|
HESX1
|
HESX homeobox 1 |
| chr2_+_216974020 | 0.09 |
ENST00000392132.2
ENST00000417391.1 |
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr3_-_147997869 | 0.09 |
ENST00000460324.1
|
RP11-501O2.4
|
RP11-501O2.4 |
| chr5_-_140700322 | 0.09 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr15_+_52155001 | 0.09 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr8_-_62602327 | 0.09 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr5_+_137203465 | 0.08 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr7_-_102985288 | 0.08 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr8_-_124749609 | 0.08 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr11_-_102651343 | 0.08 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chrX_-_15332665 | 0.08 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_+_171939240 | 0.08 |
ENST00000523513.1
|
DNM3
|
dynamin 3 |
| chr5_+_95066823 | 0.08 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr8_+_91013577 | 0.08 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr9_+_104296122 | 0.08 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr8_-_63998590 | 0.08 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr2_+_198570081 | 0.08 |
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOMEZ
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.3 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0052418 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.3 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:2000468 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0071422 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.0 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.0 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.3 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.4 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.1 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |