Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
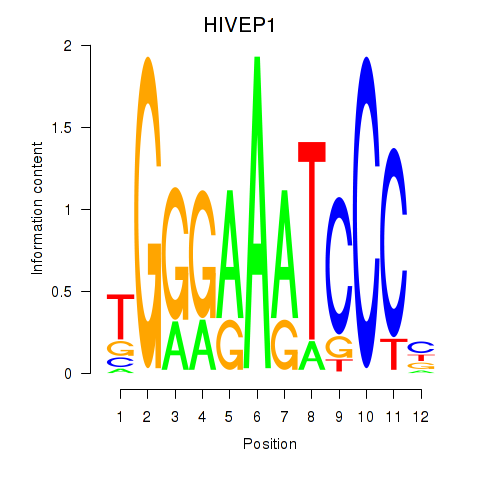
Results for HIVEP1
Z-value: 1.00
Transcription factors associated with HIVEP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIVEP1
|
ENSG00000095951.12 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIVEP1 | hg19_v2_chr6_+_12008986_12009016 | -0.43 | 3.9e-01 | Click! |
Activity profile of HIVEP1 motif
Sorted Z-values of HIVEP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_54135310 | 0.73 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr14_+_38065052 | 0.53 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_-_106697974 | 0.51 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr11_+_19799327 | 0.46 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr3_+_119814070 | 0.46 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr19_-_46272462 | 0.41 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr18_-_2982869 | 0.39 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr9_+_97562440 | 0.39 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr20_+_1115821 | 0.37 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr9_-_130477912 | 0.36 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr11_-_65314905 | 0.35 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr11_+_64052294 | 0.32 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr2_-_26569611 | 0.32 |
ENST00000541401.1
ENST00000433584.1 ENST00000333478.6 |
GPR113
|
G protein-coupled receptor 113 |
| chr8_+_94752349 | 0.32 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr4_+_4861385 | 0.32 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr14_-_69262789 | 0.32 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_+_64053005 | 0.32 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr15_+_76352178 | 0.31 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr3_-_49893907 | 0.31 |
ENST00000482582.1
|
TRAIP
|
TRAF interacting protein |
| chr3_+_187086120 | 0.31 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr11_+_19798964 | 0.31 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr5_-_150460914 | 0.30 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr17_+_40440481 | 0.30 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr1_+_1846519 | 0.30 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr14_-_69262947 | 0.29 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr16_-_4303767 | 0.29 |
ENST00000573268.1
ENST00000573042.1 |
RP11-95P2.1
|
RP11-95P2.1 |
| chr1_+_156611900 | 0.29 |
ENST00000457777.2
ENST00000424639.1 |
BCAN
|
brevican |
| chr19_+_37742773 | 0.29 |
ENST00000438770.2
ENST00000591116.1 ENST00000592712.1 |
AC012309.5
|
AC012309.5 |
| chr16_+_56970567 | 0.29 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr2_-_220252068 | 0.29 |
ENST00000430206.1
ENST00000429013.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr1_+_156611704 | 0.29 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr14_-_69446034 | 0.28 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr1_+_44457441 | 0.28 |
ENST00000466180.1
|
CCDC24
|
coiled-coil domain containing 24 |
| chr12_-_62997214 | 0.28 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr10_-_81203972 | 0.28 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr11_+_73000449 | 0.27 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr2_-_220252603 | 0.27 |
ENST00000322176.7
ENST00000273075.4 |
DNPEP
|
aspartyl aminopeptidase |
| chr15_-_41522889 | 0.27 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr17_+_37793318 | 0.27 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr11_+_64052266 | 0.27 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr20_+_327668 | 0.27 |
ENST00000382291.3
ENST00000609504.1 ENST00000382285.2 |
NRSN2
|
neurensin 2 |
| chr1_+_212738676 | 0.27 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr12_-_9913489 | 0.27 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr7_+_100271446 | 0.26 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr2_+_97481974 | 0.26 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr12_-_63328817 | 0.26 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr9_+_127054217 | 0.26 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr17_-_78428487 | 0.26 |
ENST00000562672.2
|
CTD-2526A2.2
|
CTD-2526A2.2 |
| chr1_-_94079648 | 0.26 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_160231451 | 0.26 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chrX_-_48858667 | 0.26 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr4_+_139936905 | 0.26 |
ENST00000280614.2
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
| chr11_+_47608198 | 0.26 |
ENST00000356737.2
ENST00000538490.1 |
FAM180B
|
family with sequence similarity 180, member B |
| chr20_+_327413 | 0.25 |
ENST00000609179.1
|
NRSN2
|
neurensin 2 |
| chr7_+_100271355 | 0.25 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_+_127539425 | 0.25 |
ENST00000331715.9
|
OLFML2A
|
olfactomedin-like 2A |
| chr15_+_80215113 | 0.25 |
ENST00000560255.1
|
C15orf37
|
chromosome 15 open reading frame 37 |
| chr4_+_5053162 | 0.25 |
ENST00000282908.5
|
STK32B
|
serine/threonine kinase 32B |
| chr2_-_27435634 | 0.25 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr11_+_64052454 | 0.25 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr12_+_10365082 | 0.24 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr7_-_99698338 | 0.24 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr11_-_67276100 | 0.23 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr2_-_220252530 | 0.23 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr11_+_118478313 | 0.23 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr22_-_39151434 | 0.23 |
ENST00000439339.1
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr12_+_110011571 | 0.23 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr7_+_69064300 | 0.23 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr3_+_159481464 | 0.22 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr12_+_10365404 | 0.22 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr3_+_9773409 | 0.22 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr6_-_43484718 | 0.22 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3 |
| chr5_+_140739537 | 0.22 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr11_+_64052944 | 0.22 |
ENST00000535675.1
ENST00000543383.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr19_-_39390440 | 0.22 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr12_+_57623477 | 0.22 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr14_-_69262916 | 0.21 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr20_-_46414183 | 0.21 |
ENST00000437955.1
|
SULF2
|
sulfatase 2 |
| chr14_+_105266933 | 0.21 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr19_-_4182530 | 0.21 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr7_-_74867509 | 0.21 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr17_-_79917645 | 0.21 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr7_-_149158187 | 0.21 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr4_-_122085469 | 0.21 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_-_40041925 | 0.21 |
ENST00000372862.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr17_+_6918354 | 0.21 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr14_-_69445968 | 0.21 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr1_-_159893507 | 0.21 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr17_+_37793378 | 0.21 |
ENST00000544210.2
ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr1_+_230193521 | 0.21 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr17_+_6918064 | 0.20 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr19_-_5978090 | 0.20 |
ENST00000592621.1
ENST00000034275.8 ENST00000591092.1 ENST00000591333.1 ENST00000590623.1 ENST00000439268.2 ENST00000587159.1 |
RANBP3
|
RAN binding protein 3 |
| chr6_+_149539053 | 0.20 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr16_+_4666475 | 0.20 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr17_+_40440094 | 0.20 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr15_+_40731920 | 0.20 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr3_-_9291063 | 0.20 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr3_-_49131013 | 0.20 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr22_-_19512893 | 0.20 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr19_+_41256764 | 0.20 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr16_+_85936295 | 0.20 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr15_+_85923856 | 0.19 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr19_+_45147313 | 0.19 |
ENST00000406449.4
|
PVR
|
poliovirus receptor |
| chr3_-_156878482 | 0.19 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr10_-_118031778 | 0.19 |
ENST00000369236.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr1_-_40042073 | 0.19 |
ENST00000372858.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr6_+_33176265 | 0.19 |
ENST00000374656.4
|
RING1
|
ring finger protein 1 |
| chr19_-_5785630 | 0.19 |
ENST00000586012.1
ENST00000590343.1 |
CTB-54O9.9
DUS3L
|
Uncharacterized protein dihydrouridine synthase 3-like (S. cerevisiae) |
| chr19_+_41869894 | 0.19 |
ENST00000413014.2
|
TMEM91
|
transmembrane protein 91 |
| chr19_-_46366392 | 0.19 |
ENST00000598059.1
ENST00000594293.1 ENST00000245934.7 |
SYMPK
|
symplekin |
| chr15_+_75315896 | 0.19 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr19_-_39390350 | 0.19 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr9_-_116837249 | 0.19 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr15_-_41408409 | 0.19 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr3_-_49142178 | 0.19 |
ENST00000452739.1
ENST00000414533.1 ENST00000417025.1 |
QARS
|
glutaminyl-tRNA synthetase |
| chr17_-_46623441 | 0.19 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr15_+_91411810 | 0.19 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_+_35485682 | 0.18 |
ENST00000599564.1
|
GRAMD1A
|
GRAM domain containing 1A |
| chr19_-_55628927 | 0.18 |
ENST00000263433.3
ENST00000376393.2 |
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C |
| chr17_+_21191341 | 0.18 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr9_-_136344197 | 0.18 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_-_206950996 | 0.18 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr6_-_44281043 | 0.18 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr17_-_43209862 | 0.18 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr19_-_41870026 | 0.18 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr17_+_6918093 | 0.18 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr8_+_42128812 | 0.18 |
ENST00000520810.1
ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr19_-_55770311 | 0.18 |
ENST00000412770.2
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1 |
| chr11_+_72281681 | 0.18 |
ENST00000450804.3
|
RP11-169D4.1
|
RP11-169D4.1 |
| chr1_+_110453203 | 0.18 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr19_+_18544045 | 0.18 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chrX_+_48660107 | 0.17 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr15_+_74509530 | 0.17 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr1_-_155959853 | 0.17 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_129592700 | 0.17 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr1_-_1677358 | 0.17 |
ENST00000355439.2
ENST00000400924.1 ENST00000246421.4 |
SLC35E2
|
solute carrier family 35, member E2 |
| chr7_-_22259845 | 0.17 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_53662073 | 0.17 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr1_-_36851475 | 0.17 |
ENST00000373129.3
|
STK40
|
serine/threonine kinase 40 |
| chr1_+_46640750 | 0.17 |
ENST00000372003.1
|
TSPAN1
|
tetraspanin 1 |
| chr20_-_2821756 | 0.17 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr5_+_142286887 | 0.17 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr5_-_137090028 | 0.17 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr14_-_69445793 | 0.17 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr12_+_53662110 | 0.17 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr6_+_33359582 | 0.16 |
ENST00000450504.1
|
KIFC1
|
kinesin family member C1 |
| chr9_-_131486367 | 0.16 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr19_+_17326191 | 0.16 |
ENST00000595101.1
ENST00000596136.1 ENST00000379776.4 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr16_+_75032901 | 0.16 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr1_+_38259540 | 0.16 |
ENST00000397631.3
|
MANEAL
|
mannosidase, endo-alpha-like |
| chr6_-_43484621 | 0.16 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr12_+_57624085 | 0.16 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_-_26251481 | 0.16 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr11_+_118754475 | 0.16 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr17_-_41984835 | 0.16 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr17_-_1389228 | 0.16 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr1_+_154300217 | 0.16 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr17_-_18908040 | 0.16 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr1_+_114522049 | 0.16 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr6_+_116782527 | 0.16 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chrX_+_48554986 | 0.16 |
ENST00000376687.3
ENST00000453214.2 |
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr19_+_45504688 | 0.16 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr6_-_13328050 | 0.15 |
ENST00000420456.1
|
TBC1D7
|
TBC1 domain family, member 7 |
| chr1_+_110453514 | 0.15 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr19_+_39390587 | 0.15 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr15_+_73976715 | 0.15 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr22_-_29949680 | 0.15 |
ENST00000397873.2
ENST00000490103.1 |
THOC5
|
THO complex 5 |
| chr7_+_44084262 | 0.15 |
ENST00000456905.1
ENST00000440166.1 ENST00000452943.1 ENST00000468694.1 ENST00000494774.1 ENST00000490734.2 |
DBNL
|
drebrin-like |
| chr2_-_157189180 | 0.15 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr22_-_29949657 | 0.15 |
ENST00000428374.1
|
THOC5
|
THO complex 5 |
| chr22_-_29949634 | 0.15 |
ENST00000397872.1
ENST00000397871.1 ENST00000440771.1 |
THOC5
|
THO complex 5 |
| chr1_-_156918806 | 0.15 |
ENST00000315174.8
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_+_98482175 | 0.15 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chrX_-_48814278 | 0.15 |
ENST00000455452.1
|
OTUD5
|
OTU domain containing 5 |
| chr3_+_9691117 | 0.15 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr1_+_67632083 | 0.15 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr6_-_13328362 | 0.15 |
ENST00000428109.1
ENST00000416436.1 |
TBC1D7
|
TBC1 domain family, member 7 |
| chr10_-_100027943 | 0.15 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr16_-_70835034 | 0.15 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr16_+_72088376 | 0.15 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr2_-_74753332 | 0.15 |
ENST00000451518.1
ENST00000404568.3 |
DQX1
|
DEAQ box RNA-dependent ATPase 1 |
| chr1_+_171750776 | 0.15 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
| chr1_+_54359854 | 0.15 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr16_+_57126428 | 0.15 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chr18_-_72920372 | 0.15 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr1_+_201979645 | 0.15 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr17_+_40610862 | 0.15 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr7_+_44084233 | 0.15 |
ENST00000448521.1
|
DBNL
|
drebrin-like |
| chr19_+_18496957 | 0.15 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr1_+_221051699 | 0.14 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr3_-_100565249 | 0.14 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr5_-_149792295 | 0.14 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr3_-_13114587 | 0.14 |
ENST00000429247.1
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr11_-_72492903 | 0.14 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr2_-_43453734 | 0.14 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr11_+_842808 | 0.14 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr11_+_64949899 | 0.14 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr14_+_103243813 | 0.14 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr17_-_27044760 | 0.14 |
ENST00000395243.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr19_-_43835582 | 0.14 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIVEP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.2 | 0.8 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) positive regulation of chondrocyte proliferation(GO:1902732) histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.4 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.4 | GO:1904139 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.8 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.5 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.2 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:1902462 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.5 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.1 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.2 | GO:0036017 | response to erythropoietin(GO:0036017) cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.2 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 0.4 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.3 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.9 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.3 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.0 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |