Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
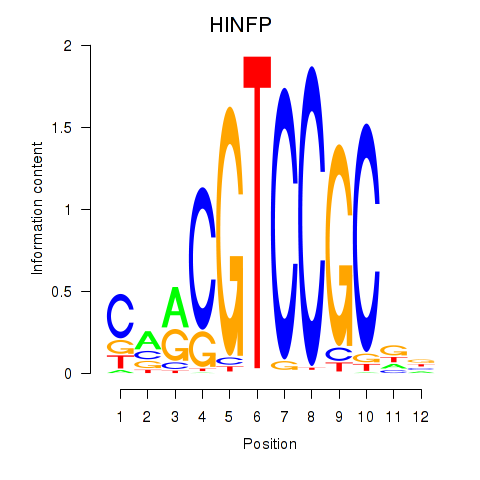
Results for HINFP
Z-value: 0.58
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | -0.98 | 3.6e-04 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_69633407 | 0.32 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr5_+_61602236 | 0.27 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr2_-_37384175 | 0.25 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr19_+_18077881 | 0.24 |
ENST00000609922.1
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr4_+_148402069 | 0.24 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chr9_+_100174344 | 0.22 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr12_+_49761147 | 0.21 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr18_-_268019 | 0.21 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr9_+_37120536 | 0.21 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr5_+_61601965 | 0.18 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr3_-_196295437 | 0.18 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr13_-_31039375 | 0.18 |
ENST00000399494.1
|
HMGB1
|
high mobility group box 1 |
| chr17_-_73179046 | 0.18 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr17_-_73178599 | 0.17 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr1_-_36916066 | 0.17 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr9_+_100174232 | 0.17 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr3_+_196295482 | 0.15 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr5_+_61602055 | 0.15 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr10_+_94608245 | 0.15 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr12_-_46766577 | 0.15 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr4_+_107236692 | 0.14 |
ENST00000510207.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr12_-_90102594 | 0.14 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_+_89514402 | 0.14 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr5_+_65440032 | 0.14 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr6_+_26104104 | 0.13 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr17_+_80186908 | 0.13 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_+_43914159 | 0.13 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr17_-_6616678 | 0.12 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr1_-_92351666 | 0.12 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chrX_-_134232630 | 0.12 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr17_-_4269768 | 0.12 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr8_-_30670053 | 0.12 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chrX_-_119695279 | 0.12 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr13_-_36705425 | 0.11 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr19_+_917287 | 0.11 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr11_+_66059339 | 0.11 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chrX_-_38186775 | 0.11 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr12_+_70637192 | 0.11 |
ENST00000550160.1
ENST00000551132.1 ENST00000552915.1 ENST00000552483.1 ENST00000550641.1 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr2_+_64068116 | 0.11 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr7_-_17980091 | 0.10 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr15_-_35280426 | 0.10 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr12_+_49760639 | 0.10 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr2_+_99953816 | 0.10 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr1_-_197115818 | 0.10 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr3_-_196295385 | 0.10 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr2_+_64068074 | 0.10 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr13_+_48877895 | 0.10 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1 |
| chr12_-_117175819 | 0.10 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr13_-_31040060 | 0.10 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr3_-_57583052 | 0.10 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr1_+_204797749 | 0.10 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chrX_-_38186811 | 0.10 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr1_+_6845384 | 0.09 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr4_+_190861941 | 0.09 |
ENST00000226798.4
|
FRG1
|
FSHD region gene 1 |
| chr19_-_460996 | 0.09 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr12_+_21654714 | 0.09 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr13_+_25875785 | 0.09 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr7_-_117513540 | 0.09 |
ENST00000160373.3
|
CTTNBP2
|
cortactin binding protein 2 |
| chr6_-_89673280 | 0.09 |
ENST00000369475.3
ENST00000369485.4 ENST00000538899.1 ENST00000265607.6 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr11_+_66045634 | 0.09 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr1_+_93913713 | 0.09 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr11_-_73309112 | 0.09 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr9_+_129622904 | 0.09 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr14_-_102026643 | 0.09 |
ENST00000555882.1
ENST00000554441.1 ENST00000553729.1 ENST00000557109.1 ENST00000557532.1 ENST00000554694.1 ENST00000554735.1 ENST00000555174.1 ENST00000557661.1 |
DIO3OS
|
DIO3 opposite strand/antisense RNA (head to head) |
| chr2_-_68479614 | 0.09 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr2_+_42396574 | 0.09 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr1_-_22109682 | 0.09 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr5_-_132073111 | 0.09 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr16_+_56225248 | 0.09 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr12_+_70637494 | 0.09 |
ENST00000548159.1
ENST00000549750.1 ENST00000551043.1 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr1_-_94703118 | 0.09 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr14_+_64971438 | 0.08 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_-_46867780 | 0.08 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr8_-_66754172 | 0.08 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr17_+_30771279 | 0.08 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr19_+_45754505 | 0.08 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
| chr2_+_42396472 | 0.08 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr20_-_30458354 | 0.08 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr3_+_110790867 | 0.08 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr14_-_92572894 | 0.08 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr17_+_29158962 | 0.08 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr3_-_150481218 | 0.08 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr4_-_111119804 | 0.08 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr5_-_141257954 | 0.08 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr3_+_110790715 | 0.08 |
ENST00000319792.3
|
PVRL3
|
poliovirus receptor-related 3 |
| chr7_+_1084206 | 0.08 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chrX_+_154299753 | 0.08 |
ENST00000369459.2
ENST00000369462.1 ENST00000411985.1 ENST00000399042.1 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr4_-_111120132 | 0.08 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr16_-_54962415 | 0.08 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr11_-_2162162 | 0.08 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr1_-_51796226 | 0.08 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr8_-_142318398 | 0.08 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr5_-_142782862 | 0.08 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_-_99947121 | 0.08 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chr12_+_52400719 | 0.08 |
ENST00000293662.4
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr4_+_107236847 | 0.07 |
ENST00000358008.3
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr8_+_26240414 | 0.07 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr6_+_56819773 | 0.07 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr4_+_190861993 | 0.07 |
ENST00000524583.1
ENST00000531991.2 |
FRG1
|
FSHD region gene 1 |
| chr14_+_64971292 | 0.07 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr9_+_34458771 | 0.07 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chrX_-_15353629 | 0.07 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr9_-_74383302 | 0.07 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr2_+_28974489 | 0.07 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr12_-_90103077 | 0.07 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr7_-_140624499 | 0.07 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr13_-_99738867 | 0.07 |
ENST00000427887.1
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr1_+_185014496 | 0.07 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr2_-_97523721 | 0.07 |
ENST00000393537.4
|
ANKRD39
|
ankyrin repeat domain 39 |
| chr2_-_54087066 | 0.07 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr6_-_139695757 | 0.07 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr1_+_6845497 | 0.07 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr17_+_11501748 | 0.07 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr17_-_16395455 | 0.07 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr5_+_56469843 | 0.07 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_+_171785824 | 0.07 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr1_-_22109484 | 0.06 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr5_+_72143988 | 0.06 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr20_-_30458019 | 0.06 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr2_+_120770645 | 0.06 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr4_+_107236722 | 0.06 |
ENST00000442366.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr17_-_46690839 | 0.06 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr22_+_29168652 | 0.06 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr16_+_81069433 | 0.06 |
ENST00000299575.4
|
ATMIN
|
ATM interactor |
| chr2_-_33824382 | 0.06 |
ENST00000238823.8
|
FAM98A
|
family with sequence similarity 98, member A |
| chr2_+_120770686 | 0.06 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr14_-_81902516 | 0.06 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr17_+_48450575 | 0.06 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr5_-_41510725 | 0.06 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr11_-_72353451 | 0.06 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr9_-_94186131 | 0.06 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr2_+_30670077 | 0.06 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr12_+_107349497 | 0.05 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr7_+_29234375 | 0.05 |
ENST00000409350.1
ENST00000495789.2 ENST00000539389.1 |
CHN2
|
chimerin 2 |
| chr5_-_158636512 | 0.05 |
ENST00000424310.2
|
RNF145
|
ring finger protein 145 |
| chr8_-_100905850 | 0.05 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr2_+_120302055 | 0.05 |
ENST00000598644.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr17_-_42276574 | 0.05 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_-_40157345 | 0.05 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr3_-_113415441 | 0.05 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr10_-_62761188 | 0.05 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr4_+_108746282 | 0.05 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr2_-_73053126 | 0.05 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr2_+_157292859 | 0.05 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr12_+_12764773 | 0.05 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr13_-_48612067 | 0.05 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr22_-_43010859 | 0.05 |
ENST00000339677.6
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr13_-_22178284 | 0.05 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr12_+_56137064 | 0.05 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr8_-_100905925 | 0.05 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr5_+_56469775 | 0.05 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr10_+_94352956 | 0.05 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr12_+_12510352 | 0.05 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr12_+_58005204 | 0.05 |
ENST00000286494.4
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr19_-_35323762 | 0.04 |
ENST00000590963.1
|
CTC-523E23.4
|
CTC-523E23.4 |
| chr6_+_44355257 | 0.04 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr6_+_116692102 | 0.04 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr1_+_22351977 | 0.04 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr3_-_129612394 | 0.04 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr16_-_28074822 | 0.04 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr8_+_110656344 | 0.04 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr7_+_97910962 | 0.04 |
ENST00000539286.1
|
BRI3
|
brain protein I3 |
| chr19_+_6372444 | 0.04 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr5_+_56469939 | 0.04 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr3_-_49449350 | 0.04 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr13_+_25875662 | 0.04 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr16_+_56899114 | 0.04 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr8_-_102217796 | 0.04 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr17_+_43972010 | 0.04 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr3_-_57583130 | 0.04 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr5_-_159546396 | 0.04 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr17_+_38024417 | 0.04 |
ENST00000348931.4
ENST00000583811.1 ENST00000584588.1 ENST00000377940.3 |
ZPBP2
|
zona pellucida binding protein 2 |
| chr4_-_156298028 | 0.04 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr8_-_101733794 | 0.04 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_-_122131696 | 0.04 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr8_-_100905363 | 0.04 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr1_-_200379129 | 0.04 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr3_-_194207388 | 0.04 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chrX_+_154299690 | 0.04 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr11_+_28131821 | 0.04 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr10_+_75532028 | 0.04 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr1_+_212208919 | 0.04 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr6_-_111804905 | 0.04 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr12_+_102271436 | 0.04 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr4_-_492891 | 0.04 |
ENST00000338977.5
ENST00000511833.2 |
ZNF721
|
zinc finger protein 721 |
| chr3_-_49449521 | 0.03 |
ENST00000431929.1
ENST00000418115.1 |
RHOA
|
ras homolog family member A |
| chr9_-_115095123 | 0.03 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr1_-_169455169 | 0.03 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr2_+_120302041 | 0.03 |
ENST00000442513.3
ENST00000413369.3 |
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr2_-_170219037 | 0.03 |
ENST00000443831.1
|
LRP2
|
low density lipoprotein receptor-related protein 2 |
| chr18_+_67068228 | 0.03 |
ENST00000382713.5
|
DOK6
|
docking protein 6 |
| chr4_-_6202247 | 0.03 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr11_+_47586982 | 0.03 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr12_+_72666407 | 0.03 |
ENST00000261180.4
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr4_-_107237374 | 0.03 |
ENST00000361687.4
ENST00000507696.1 ENST00000394708.2 ENST00000509532.1 |
TBCK
|
TBC1 domain containing kinase |
| chr4_-_6202291 | 0.03 |
ENST00000282924.5
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr1_-_16563641 | 0.03 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr1_-_92351769 | 0.03 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr1_-_200379180 | 0.03 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr12_+_12509990 | 0.03 |
ENST00000542728.1
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0034699 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.3 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |