Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HDX
Z-value: 0.15
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HDX | hg19_v2_chrX_-_83757399_83757487 | 0.75 | 8.3e-02 | Click! |
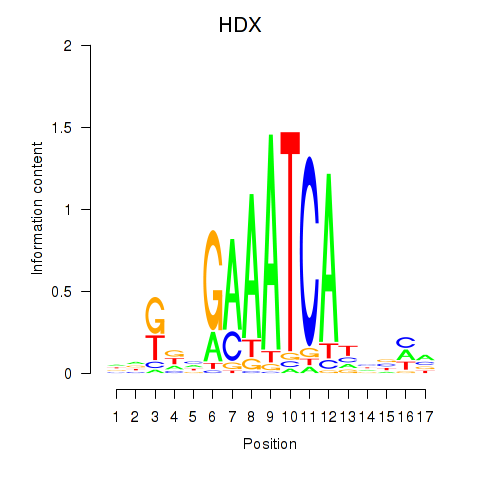
Activity profile of HDX motif
Sorted Z-values of HDX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113594279 | 0.10 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr2_+_113735575 | 0.09 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr7_-_139756791 | 0.08 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr13_+_97928395 | 0.07 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr5_-_135290651 | 0.06 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr2_+_102758210 | 0.06 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr1_+_84629976 | 0.06 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_-_21186144 | 0.06 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr2_+_102758271 | 0.05 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr15_-_54025300 | 0.05 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr3_-_165555200 | 0.04 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chrX_+_100224676 | 0.04 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr14_+_79745682 | 0.03 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr1_+_84630053 | 0.03 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_+_48807155 | 0.03 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr3_+_107241882 | 0.03 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr2_+_128403720 | 0.03 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr1_+_84630352 | 0.02 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_+_1877204 | 0.02 |
ENST00000427358.2
|
FAHD1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr2_+_128403439 | 0.02 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr1_-_11024258 | 0.02 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr3_-_51533966 | 0.02 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr1_-_48866517 | 0.02 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr7_-_107883678 | 0.01 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr14_+_71788096 | 0.01 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr5_+_140165876 | 0.01 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr6_+_144471643 | 0.01 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr12_-_31743901 | 0.01 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr2_+_149447783 | 0.01 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr5_-_35230649 | 0.01 |
ENST00000382002.5
|
PRLR
|
prolactin receptor |
| chr5_+_140625147 | 0.01 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr12_-_39734783 | 0.01 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chrX_+_27608490 | 0.01 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr6_-_116381918 | 0.01 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr14_+_79746249 | 0.01 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr11_+_9685604 | 0.01 |
ENST00000447399.2
ENST00000318950.6 |
SWAP70
|
SWAP switching B-cell complex 70kDa subunit |
| chr2_+_169926047 | 0.00 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_169680745 | 0.00 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr3_+_54157480 | 0.00 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr19_+_36602104 | 0.00 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0035234 | smooth muscle adaptation(GO:0014805) ectopic germ cell programmed cell death(GO:0035234) negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |