Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
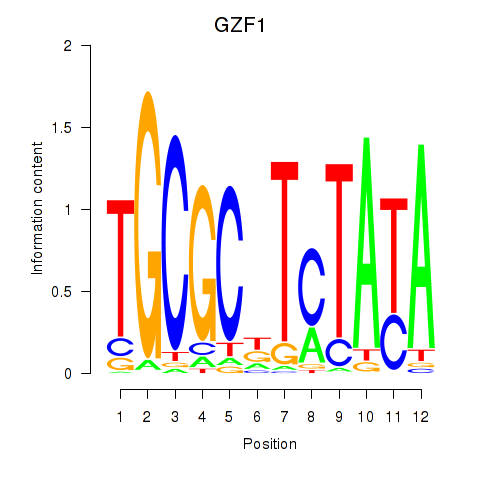
Results for GZF1
Z-value: 0.76
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.11 | GDNF inducible zinc finger protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GZF1 | hg19_v2_chr20_+_23342783_23342957 | -0.16 | 7.6e-01 | Click! |
Activity profile of GZF1 motif
Sorted Z-values of GZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_79115503 | 1.30 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr12_-_107168696 | 0.62 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr12_-_76478386 | 0.45 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_-_114035026 | 0.45 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr4_-_147443043 | 0.43 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr3_-_170588163 | 0.38 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr4_-_89619386 | 0.37 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr12_+_102513950 | 0.37 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr14_-_92413727 | 0.34 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr10_+_89264625 | 0.34 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr2_-_74776586 | 0.33 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr2_-_150444300 | 0.32 |
ENST00000303319.5
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr4_-_147442817 | 0.32 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr19_-_44384291 | 0.31 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr12_-_76478417 | 0.30 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr10_-_126849626 | 0.30 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr16_-_30582888 | 0.30 |
ENST00000563707.1
ENST00000567855.1 |
ZNF688
|
zinc finger protein 688 |
| chr3_-_87039662 | 0.29 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr14_-_92413353 | 0.28 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr12_-_76478446 | 0.27 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_+_104458235 | 0.26 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr2_-_150444116 | 0.26 |
ENST00000428879.1
ENST00000422782.2 |
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr1_-_11107280 | 0.25 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr19_+_36157715 | 0.20 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr12_-_102513843 | 0.20 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr13_+_98086445 | 0.19 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr9_-_124976185 | 0.19 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr5_+_170814803 | 0.16 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr7_-_105029329 | 0.16 |
ENST00000393651.3
ENST00000460391.1 |
SRPK2
|
SRSF protein kinase 2 |
| chr7_-_27183263 | 0.15 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr10_+_97759848 | 0.15 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr14_-_52535712 | 0.13 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr3_+_30647994 | 0.13 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr16_-_46797149 | 0.12 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr4_+_26344754 | 0.12 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_+_60028818 | 0.12 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr3_+_30648066 | 0.10 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr6_+_30525051 | 0.09 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr16_+_86612112 | 0.09 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr11_-_17498348 | 0.09 |
ENST00000389817.3
ENST00000302539.4 |
ABCC8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr10_+_14880364 | 0.09 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr14_-_53619816 | 0.08 |
ENST00000323669.5
ENST00000395606.1 ENST00000357758.3 |
DDHD1
|
DDHD domain containing 1 |
| chr7_-_26904317 | 0.08 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr6_+_30524663 | 0.08 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr1_+_26606608 | 0.08 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr3_-_87040233 | 0.08 |
ENST00000398399.2
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr9_-_124976154 | 0.07 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chrX_+_54835493 | 0.05 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr5_-_176827631 | 0.05 |
ENST00000358571.2
|
PFN3
|
profilin 3 |
| chrX_+_48367338 | 0.05 |
ENST00000359882.4
ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN
|
porcupine homolog (Drosophila) |
| chr16_+_87636474 | 0.04 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr1_-_144995074 | 0.03 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_+_107168418 | 0.03 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr6_-_30524951 | 0.03 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr19_+_38880695 | 0.03 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_+_64937625 | 0.03 |
ENST00000377185.2
|
SPDYC
|
speedy/RINGO cell cycle regulator family member C |
| chr3_-_79816965 | 0.02 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr11_+_101918153 | 0.02 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr1_-_226374373 | 0.02 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr10_-_14880566 | 0.02 |
ENST00000378442.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr1_+_41174988 | 0.01 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr4_-_147442982 | 0.01 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr20_-_31331212 | 0.01 |
ENST00000474815.2
ENST00000446419.2 |
COMMD7
|
COMM domain containing 7 |
| chr10_-_126849588 | 0.00 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GZF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.0 | 0.3 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |