Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
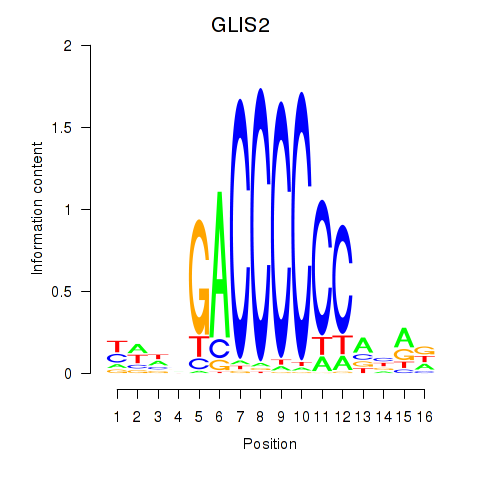
Results for GLIS2
Z-value: 1.45
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg19_v2_chr16_+_4382225_4382225 | -0.35 | 5.0e-01 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_615942 | 2.47 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr11_-_615570 | 2.11 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr17_+_41158742 | 1.57 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr19_-_55669093 | 1.34 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr20_-_62203808 | 1.23 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr19_+_55795493 | 1.23 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr1_-_156675564 | 0.90 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_+_41177220 | 0.86 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr11_-_627143 | 0.65 |
ENST00000176195.3
|
SCT
|
secretin |
| chr5_+_172484377 | 0.64 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chr2_+_24272543 | 0.63 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr1_-_27701307 | 0.63 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr6_-_41673552 | 0.63 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr1_-_156675535 | 0.59 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_-_156675368 | 0.58 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr20_+_52824367 | 0.58 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr2_-_27718052 | 0.57 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr19_-_6720686 | 0.54 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_-_821931 | 0.54 |
ENST00000359894.2
ENST00000520876.3 ENST00000519502.1 |
LPPR3
|
hsa-mir-3187 |
| chr3_-_52719888 | 0.53 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr1_-_186649543 | 0.52 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_+_24272576 | 0.52 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr1_-_54411255 | 0.48 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr6_+_131571535 | 0.47 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr13_-_20767037 | 0.46 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr5_+_140566 | 0.46 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr9_+_139839686 | 0.45 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr7_+_77167343 | 0.45 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_-_55668093 | 0.44 |
ENST00000588882.1
ENST00000586858.1 |
TNNI3
|
troponin I type 3 (cardiac) |
| chr9_+_139847347 | 0.42 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr8_-_103136481 | 0.41 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chr4_-_111120334 | 0.39 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr7_+_77167376 | 0.39 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_+_36602104 | 0.38 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr17_-_41132088 | 0.38 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr19_-_51071302 | 0.38 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr1_-_31661000 | 0.38 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr12_-_120907374 | 0.37 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr2_+_170655789 | 0.37 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr19_-_50979981 | 0.37 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr14_-_24615523 | 0.36 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr21_-_45671014 | 0.36 |
ENST00000436357.1
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr5_-_131826457 | 0.36 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr2_+_48757278 | 0.36 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chrX_+_53449805 | 0.35 |
ENST00000414955.2
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chrX_+_53449839 | 0.34 |
ENST00000457095.1
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr20_-_30310797 | 0.34 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr19_+_15619299 | 0.33 |
ENST00000269703.3
|
CYP4F22
|
cytochrome P450, family 4, subfamily F, polypeptide 22 |
| chr13_-_108867846 | 0.32 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr12_-_95942563 | 0.32 |
ENST00000549639.1
ENST00000551837.1 |
USP44
|
ubiquitin specific peptidase 44 |
| chr3_+_118892411 | 0.32 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr16_-_29910365 | 0.32 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr11_-_1619524 | 0.32 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr11_-_63684316 | 0.31 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr1_-_54411240 | 0.30 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_-_115632035 | 0.30 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr5_+_68788594 | 0.30 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr19_+_56159421 | 0.30 |
ENST00000587213.1
|
CCDC106
|
coiled-coil domain containing 106 |
| chr19_+_17858509 | 0.30 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr14_-_54955376 | 0.30 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr15_+_90234028 | 0.30 |
ENST00000268130.7
ENST00000560294.1 ENST00000558000.1 |
WDR93
|
WD repeat domain 93 |
| chr7_+_133812052 | 0.30 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr6_-_30815936 | 0.29 |
ENST00000442852.1
|
XXbac-BPG27H4.8
|
XXbac-BPG27H4.8 |
| chr19_+_45417504 | 0.27 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr19_+_45349630 | 0.27 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr14_-_24615805 | 0.27 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr14_-_105635090 | 0.27 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr11_-_3013482 | 0.27 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr17_-_7120525 | 0.27 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr1_-_6550625 | 0.26 |
ENST00000377725.1
ENST00000340850.5 |
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr19_-_35454953 | 0.26 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr2_+_187350883 | 0.26 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr17_+_48503603 | 0.26 |
ENST00000502667.1
|
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr5_+_169011033 | 0.26 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr11_-_3013497 | 0.26 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr6_+_144164455 | 0.25 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr13_-_108867101 | 0.25 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr19_+_56159362 | 0.24 |
ENST00000593069.1
ENST00000308964.3 |
CCDC106
|
coiled-coil domain containing 106 |
| chr2_+_71357744 | 0.24 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chrX_+_48660565 | 0.24 |
ENST00000413163.2
ENST00000441703.1 ENST00000426196.1 |
HDAC6
|
histone deacetylase 6 |
| chr3_-_63849571 | 0.24 |
ENST00000295899.5
|
THOC7
|
THO complex 7 homolog (Drosophila) |
| chr19_+_45349432 | 0.24 |
ENST00000252485.4
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr17_-_45266542 | 0.24 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr8_+_27182862 | 0.24 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr19_+_56159509 | 0.24 |
ENST00000586790.1
ENST00000591578.1 ENST00000588740.1 |
CCDC106
|
coiled-coil domain containing 106 |
| chr8_-_143833918 | 0.24 |
ENST00000359228.3
|
LYPD2
|
LY6/PLAUR domain containing 2 |
| chr5_+_169659950 | 0.24 |
ENST00000593851.1
|
C5orf58
|
chromosome 5 open reading frame 58 |
| chr1_-_151138422 | 0.24 |
ENST00000440902.2
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr1_+_43424698 | 0.23 |
ENST00000431759.1
|
SLC2A1-AS1
|
SLC2A1 antisense RNA 1 |
| chr11_-_3013316 | 0.23 |
ENST00000430811.1
|
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr4_-_111120132 | 0.23 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr17_+_48503519 | 0.23 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr11_-_117688216 | 0.23 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr13_-_95248511 | 0.23 |
ENST00000261296.5
|
TGDS
|
TDP-glucose 4,6-dehydratase |
| chr19_+_45417812 | 0.22 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr2_-_113999260 | 0.22 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr8_+_27183033 | 0.22 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr17_+_30348024 | 0.22 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr4_-_170679024 | 0.22 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr6_+_43044003 | 0.21 |
ENST00000230419.4
ENST00000476760.1 ENST00000471863.1 ENST00000349241.2 ENST00000352931.2 ENST00000345201.2 |
PTK7
|
protein tyrosine kinase 7 |
| chr22_-_30661807 | 0.21 |
ENST00000403389.1
|
OSM
|
oncostatin M |
| chr2_-_157186630 | 0.21 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr19_-_3772209 | 0.21 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr3_+_151531859 | 0.21 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr19_+_45251804 | 0.21 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr20_-_30310693 | 0.21 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chrX_+_53449887 | 0.21 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr9_+_135285579 | 0.20 |
ENST00000343036.2
ENST00000393216.2 |
C9orf171
|
chromosome 9 open reading frame 171 |
| chr4_-_106394866 | 0.20 |
ENST00000502596.1
|
PPA2
|
pyrophosphatase (inorganic) 2 |
| chr10_+_115438920 | 0.20 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr18_+_56531584 | 0.20 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr17_-_4463856 | 0.20 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr14_-_21493884 | 0.20 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr14_+_103566481 | 0.20 |
ENST00000380069.3
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr8_-_104427289 | 0.20 |
ENST00000543107.1
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr15_+_63796779 | 0.19 |
ENST00000561442.1
ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3
|
ubiquitin specific peptidase 3 |
| chr20_-_30310656 | 0.19 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr6_-_117086873 | 0.19 |
ENST00000368557.4
|
FAM162B
|
family with sequence similarity 162, member B |
| chr17_-_79166176 | 0.19 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr14_-_50999307 | 0.19 |
ENST00000013125.4
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_114292449 | 0.19 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr19_+_45417921 | 0.19 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr22_+_22681656 | 0.19 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr19_-_2282167 | 0.19 |
ENST00000342063.3
|
C19orf35
|
chromosome 19 open reading frame 35 |
| chr3_-_49933249 | 0.18 |
ENST00000434765.1
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr6_-_114292284 | 0.18 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr11_+_61197508 | 0.18 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr8_-_95565673 | 0.18 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr5_+_102090974 | 0.18 |
ENST00000512073.1
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_102631844 | 0.18 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr1_+_91966384 | 0.18 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chrX_-_48823165 | 0.18 |
ENST00000419374.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr19_-_14586125 | 0.17 |
ENST00000292513.3
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa |
| chr14_-_92302784 | 0.17 |
ENST00000340892.5
ENST00000360594.5 |
TC2N
|
tandem C2 domains, nuclear |
| chr12_-_51422017 | 0.17 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr19_-_17799008 | 0.17 |
ENST00000519716.2
|
UNC13A
|
unc-13 homolog A (C. elegans) |
| chr12_+_56401268 | 0.17 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr8_+_38243967 | 0.17 |
ENST00000524874.1
ENST00000379957.4 ENST00000523983.2 |
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr11_-_111637083 | 0.17 |
ENST00000427203.2
ENST00000341980.6 ENST00000311129.5 ENST00000393055.2 ENST00000426998.2 ENST00000527614.1 |
PPP2R1B
|
protein phosphatase 2, regulatory subunit A, beta |
| chr16_-_29910853 | 0.17 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr12_-_95467267 | 0.17 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr14_+_101293687 | 0.17 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr12_-_95942613 | 0.17 |
ENST00000393091.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr12_+_50451462 | 0.17 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr1_-_156647189 | 0.16 |
ENST00000368223.3
|
NES
|
nestin |
| chr19_+_7413835 | 0.16 |
ENST00000576789.1
|
CTB-133G6.1
|
CTB-133G6.1 |
| chr20_-_30458019 | 0.16 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr3_+_111578131 | 0.16 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_-_95467356 | 0.16 |
ENST00000393101.3
ENST00000333003.5 |
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr17_-_40169429 | 0.16 |
ENST00000316603.7
ENST00000588641.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr18_+_12948000 | 0.16 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr17_+_18012020 | 0.15 |
ENST00000205890.5
|
MYO15A
|
myosin XVA |
| chr3_-_49933506 | 0.15 |
ENST00000440292.1
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr11_-_46142615 | 0.15 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr1_-_40157345 | 0.15 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr17_-_4269768 | 0.15 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr1_+_64936428 | 0.15 |
ENST00000371073.2
ENST00000290039.5 |
CACHD1
|
cache domain containing 1 |
| chr19_-_17185848 | 0.15 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr1_+_150954493 | 0.15 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr6_-_153304697 | 0.15 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr2_+_29033682 | 0.15 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr2_-_220197351 | 0.15 |
ENST00000392083.1
|
RESP18
|
regulated endocrine-specific protein 18 |
| chr1_+_87595497 | 0.15 |
ENST00000471417.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr3_+_52719936 | 0.15 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr15_+_52311398 | 0.15 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr11_-_14665163 | 0.15 |
ENST00000418988.2
|
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr17_-_40169161 | 0.14 |
ENST00000589586.2
ENST00000426588.3 ENST00000589576.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr8_+_38243951 | 0.14 |
ENST00000297720.5
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_+_149865377 | 0.14 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_+_181845074 | 0.14 |
ENST00000602959.1
ENST00000602479.1 ENST00000392415.2 ENST00000602291.1 |
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr2_+_89952792 | 0.14 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr22_-_38794490 | 0.14 |
ENST00000400206.2
|
CSNK1E
|
casein kinase 1, epsilon |
| chr17_-_36760865 | 0.14 |
ENST00000584266.1
|
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr12_-_51785182 | 0.14 |
ENST00000356317.3
ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr8_+_38243821 | 0.14 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr7_-_158380371 | 0.14 |
ENST00000389418.4
ENST00000389416.4 |
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr22_-_50700140 | 0.14 |
ENST00000215659.8
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr6_-_153304148 | 0.13 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr22_-_30685596 | 0.13 |
ENST00000404953.3
ENST00000407689.3 |
GATSL3
|
GATS protein-like 3 |
| chr10_+_94608245 | 0.13 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr6_-_32908792 | 0.13 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr3_-_128840604 | 0.13 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr17_-_79623597 | 0.13 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr16_+_2198604 | 0.13 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr14_-_21493649 | 0.13 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr3_+_151531810 | 0.13 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr8_+_118533049 | 0.13 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr2_+_37571717 | 0.13 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr19_-_2702681 | 0.13 |
ENST00000382159.3
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr1_+_43637996 | 0.13 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr1_-_22215192 | 0.13 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr1_+_65613217 | 0.13 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr3_-_43663519 | 0.12 |
ENST00000427171.1
ENST00000292246.3 |
ANO10
|
anoctamin 10 |
| chr18_-_19180681 | 0.12 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr3_+_180630444 | 0.12 |
ENST00000491062.1
ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr12_+_50017327 | 0.12 |
ENST00000261897.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr19_-_39108568 | 0.12 |
ENST00000586296.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr17_+_64831228 | 0.12 |
ENST00000533854.1
|
CACNG5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr1_+_47901689 | 0.12 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr7_+_130794846 | 0.12 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 0.6 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 0.6 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.2 | 0.5 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 2.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.5 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.1 | GO:0071106 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.5 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.7 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.5 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.1 | 0.3 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:1903911 | positive regulation of receptor clustering(GO:1903911) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.3 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.3 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.6 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 1.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 1.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 2.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.5 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.3 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 0.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 1.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.6 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 4.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 5.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |