Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
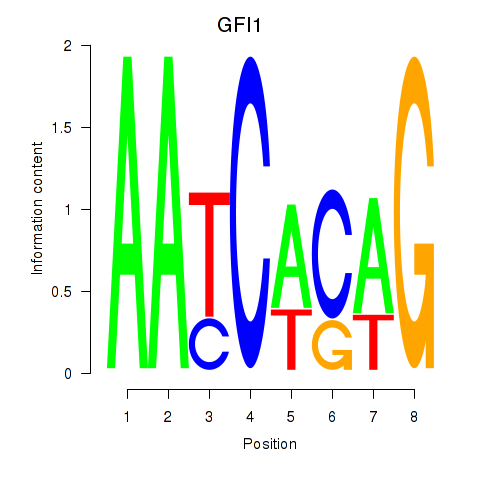
Results for GFI1
Z-value: 0.70
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | growth factor independent 1 transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92952433_92952489 | 0.21 | 6.9e-01 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_47448102 | 0.72 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr12_-_106697974 | 0.43 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chrX_+_53078465 | 0.42 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr15_+_96875657 | 0.38 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_+_45908974 | 0.36 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr3_-_52002403 | 0.35 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr17_+_79495397 | 0.35 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr8_-_145642203 | 0.34 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chrX_+_103294483 | 0.31 |
ENST00000355016.3
|
H2BFM
|
H2B histone family, member M |
| chr3_-_33686925 | 0.30 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_+_3540043 | 0.28 |
ENST00000574218.1
|
CTNS
|
cystinosin, lysosomal cystine transporter |
| chr12_+_56732658 | 0.28 |
ENST00000228534.4
|
IL23A
|
interleukin 23, alpha subunit p19 |
| chr9_-_85882145 | 0.28 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr4_-_119759795 | 0.28 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr18_-_46895066 | 0.28 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr13_-_30951282 | 0.27 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr12_-_21910775 | 0.27 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr19_+_44617511 | 0.27 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr1_+_62439037 | 0.26 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr16_+_21623958 | 0.26 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr15_-_44069513 | 0.26 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr7_+_120591170 | 0.25 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr2_-_61389240 | 0.24 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr17_+_80317121 | 0.24 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr11_-_10920714 | 0.23 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr7_+_23637118 | 0.23 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chrX_+_53449887 | 0.23 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr19_+_41281060 | 0.22 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr1_-_67142710 | 0.22 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr3_+_190281229 | 0.22 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr17_-_57229155 | 0.21 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_+_121570631 | 0.21 |
ENST00000546057.1
ENST00000377162.2 ENST00000328963.5 ENST00000535250.1 ENST00000541446.1 |
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr19_-_58204128 | 0.21 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr1_+_162336686 | 0.20 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr19_-_51875894 | 0.20 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr6_-_32977345 | 0.19 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr8_+_118532937 | 0.19 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chr7_-_20826504 | 0.19 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chrX_-_54069576 | 0.19 |
ENST00000445025.1
ENST00000322659.8 |
PHF8
|
PHD finger protein 8 |
| chr15_+_67458861 | 0.19 |
ENST00000558428.1
ENST00000558827.1 |
SMAD3
|
SMAD family member 3 |
| chr9_-_127710292 | 0.19 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr7_+_114055052 | 0.18 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr11_-_82745238 | 0.18 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr8_+_132320792 | 0.18 |
ENST00000519695.1
ENST00000524275.1 |
CTD-2501M5.1
|
CTD-2501M5.1 |
| chr13_-_33780133 | 0.17 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr2_-_61389168 | 0.17 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr1_-_179834311 | 0.17 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr15_+_84116106 | 0.17 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr2_+_27799389 | 0.16 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr16_+_2533020 | 0.16 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr11_-_26743546 | 0.16 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr6_-_138820624 | 0.16 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr11_+_5646213 | 0.15 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr1_-_242612779 | 0.15 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr19_+_50353944 | 0.15 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr6_-_56492816 | 0.15 |
ENST00000522360.1
|
DST
|
dystonin |
| chr13_+_76362974 | 0.15 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr8_-_133772870 | 0.15 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr1_-_54879140 | 0.15 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr3_-_195619579 | 0.15 |
ENST00000428187.1
|
TNK2
|
tyrosine kinase, non-receptor, 2 |
| chr15_+_96876340 | 0.15 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_4754339 | 0.15 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr11_-_4629388 | 0.15 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr1_-_152196669 | 0.14 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr9_+_2158485 | 0.14 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_159084188 | 0.14 |
ENST00000367081.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr6_+_99282570 | 0.14 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr12_-_31158902 | 0.14 |
ENST00000544329.1
ENST00000418254.2 ENST00000222396.5 |
RP11-551L14.4
|
RP11-551L14.4 |
| chr20_-_1569278 | 0.14 |
ENST00000262929.5
ENST00000567028.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr7_+_55980331 | 0.14 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr17_-_7218631 | 0.14 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr10_-_73497581 | 0.14 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr6_+_26440700 | 0.14 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr11_-_82746587 | 0.14 |
ENST00000528379.1
ENST00000534103.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr6_+_31949801 | 0.13 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr2_-_40657397 | 0.13 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_+_90192974 | 0.13 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr2_+_148602058 | 0.13 |
ENST00000241416.7
ENST00000535787.1 ENST00000404590.1 |
ACVR2A
|
activin A receptor, type IIA |
| chr10_-_129924468 | 0.13 |
ENST00000368653.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr21_+_38593701 | 0.13 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr19_-_3761673 | 0.13 |
ENST00000316757.3
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr19_+_50354393 | 0.12 |
ENST00000391842.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr12_+_44229846 | 0.12 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr2_+_239335636 | 0.12 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr14_-_54425475 | 0.12 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr5_+_109025067 | 0.12 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chrX_-_54069253 | 0.12 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr6_-_10412600 | 0.12 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr6_-_37225367 | 0.12 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr1_-_242612726 | 0.12 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr19_-_19302931 | 0.12 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr9_+_2158443 | 0.12 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_25279759 | 0.12 |
ENST00000377969.3
|
LRRC16A
|
leucine rich repeat containing 16A |
| chr19_+_4402659 | 0.12 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr19_+_18303992 | 0.12 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr17_+_75449889 | 0.12 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr7_+_16793160 | 0.12 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr16_+_2732476 | 0.12 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr10_-_129924611 | 0.12 |
ENST00000368654.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr6_-_27835357 | 0.12 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr1_+_12524965 | 0.12 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_+_69869237 | 0.12 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr18_+_56532100 | 0.12 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr17_-_36884451 | 0.12 |
ENST00000595377.1
|
AC006449.1
|
NS5ATP13TP1; Uncharacterized protein |
| chr12_-_118406028 | 0.12 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr19_+_36393422 | 0.12 |
ENST00000437550.2
|
HCST
|
hematopoietic cell signal transducer |
| chr17_-_45056606 | 0.12 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr20_-_62203808 | 0.12 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr5_-_150284351 | 0.12 |
ENST00000427179.1
|
ZNF300
|
zinc finger protein 300 |
| chr12_+_113587558 | 0.12 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr17_+_61699766 | 0.12 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr7_-_37382360 | 0.12 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr17_-_1395954 | 0.12 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr17_+_4675175 | 0.11 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr17_+_44790515 | 0.11 |
ENST00000576346.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr1_+_203651937 | 0.11 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr9_+_2159850 | 0.11 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_+_75450099 | 0.11 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr12_-_95510743 | 0.11 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr2_-_231989808 | 0.11 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr17_-_56595196 | 0.11 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr8_-_28965684 | 0.11 |
ENST00000523130.1
|
KIF13B
|
kinesin family member 13B |
| chr10_+_180405 | 0.11 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr17_+_5973740 | 0.11 |
ENST00000576083.1
|
WSCD1
|
WSC domain containing 1 |
| chr9_+_70917276 | 0.11 |
ENST00000342833.2
|
FOXD4L3
|
forkhead box D4-like 3 |
| chr19_-_55660561 | 0.11 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr3_-_188665428 | 0.11 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr5_+_140529630 | 0.11 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr15_+_92396920 | 0.11 |
ENST00000318445.6
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr5_+_140729649 | 0.11 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr10_-_14996017 | 0.11 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr4_+_41614720 | 0.11 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chrX_+_114874727 | 0.11 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr13_+_76413852 | 0.11 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr19_+_50354462 | 0.11 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chrX_-_73512177 | 0.11 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chrX_-_14047996 | 0.11 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr15_+_99433570 | 0.11 |
ENST00000558898.1
|
IGF1R
|
insulin-like growth factor 1 receptor |
| chr7_+_45067265 | 0.11 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr17_+_38337491 | 0.11 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr6_+_31795506 | 0.11 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr9_-_95244781 | 0.11 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr12_-_59314246 | 0.11 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr10_+_180643 | 0.11 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr17_+_78518617 | 0.11 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr4_-_467892 | 0.10 |
ENST00000506646.1
ENST00000505900.1 |
ZNF721
|
zinc finger protein 721 |
| chr10_+_47894023 | 0.10 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr2_+_101437487 | 0.10 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr12_-_4758159 | 0.10 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr2_+_238499812 | 0.10 |
ENST00000452801.1
|
AC104667.3
|
AC104667.3 |
| chr19_+_44488330 | 0.10 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr1_+_201755452 | 0.10 |
ENST00000438083.1
|
NAV1
|
neuron navigator 1 |
| chr18_+_55711575 | 0.10 |
ENST00000356462.6
ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_+_31982539 | 0.10 |
ENST00000435363.2
ENST00000425700.2 |
C4B
|
complement component 4B (Chido blood group) |
| chr5_+_59783941 | 0.10 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr9_-_70178815 | 0.10 |
ENST00000377420.1
|
FOXD4L5
|
forkhead box D4-like 5 |
| chr8_-_27115903 | 0.10 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr4_-_155533787 | 0.10 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr18_-_78005231 | 0.10 |
ENST00000470488.2
ENST00000353265.3 |
PARD6G
|
par-6 family cell polarity regulator gamma |
| chr19_-_43099070 | 0.10 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr4_+_86749045 | 0.10 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_11374904 | 0.10 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr6_+_30295036 | 0.10 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr1_+_104104379 | 0.10 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr2_-_37415690 | 0.10 |
ENST00000260637.3
ENST00000535679.1 ENST00000379149.2 |
SULT6B1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr19_-_18391708 | 0.09 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chrX_+_133507389 | 0.09 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr19_+_41281282 | 0.09 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr1_+_23695680 | 0.09 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr17_+_75450146 | 0.09 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr17_+_76311791 | 0.09 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr19_+_36393367 | 0.09 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr16_-_90038866 | 0.09 |
ENST00000314994.3
|
CENPBD1
|
CENPB DNA-binding domains containing 1 |
| chr9_-_34376851 | 0.09 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr3_+_141106458 | 0.09 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_+_1665253 | 0.09 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_+_117348742 | 0.09 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr22_+_27068704 | 0.09 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr20_+_58179582 | 0.09 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr20_+_36373032 | 0.09 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr12_+_57998400 | 0.09 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr22_-_42322795 | 0.09 |
ENST00000291232.3
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C |
| chr12_+_100867486 | 0.09 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr17_-_40761375 | 0.09 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr16_-_18441131 | 0.09 |
ENST00000339303.5
|
NPIPA8
|
nuclear pore complex interacting protein family, member A8 |
| chr6_+_37897735 | 0.09 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr17_-_62340581 | 0.09 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr11_+_1889880 | 0.09 |
ENST00000405957.2
|
LSP1
|
lymphocyte-specific protein 1 |
| chr11_-_59436453 | 0.09 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr3_+_40518599 | 0.09 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr11_+_71900703 | 0.09 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr17_-_8093471 | 0.08 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr12_+_117176113 | 0.08 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr11_-_64703354 | 0.08 |
ENST00000532246.1
ENST00000279168.2 |
GPHA2
|
glycoprotein hormone alpha 2 |
| chr1_-_150669604 | 0.08 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr17_+_40761660 | 0.08 |
ENST00000251413.3
ENST00000591509.1 |
TUBG1
|
tubulin, gamma 1 |
| chr2_+_45168875 | 0.08 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr10_-_91403625 | 0.08 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr1_+_236557569 | 0.08 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr12_-_56843161 | 0.08 |
ENST00000554616.1
ENST00000553532.1 ENST00000229201.4 |
TIMELESS
|
timeless circadian clock |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.2 | GO:0043132 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) NAD transport(GO:0043132) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.3 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.2 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0098736 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.1 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0072099 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0032639 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0038185 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.0 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |