Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
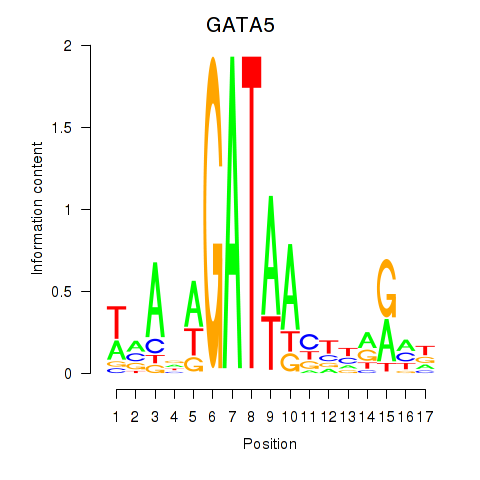
Results for GATA5
Z-value: 0.59
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.6 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA5 | hg19_v2_chr20_-_61051026_61051057 | -0.27 | 6.1e-01 | Click! |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_79115503 | 0.32 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr5_+_75904918 | 0.27 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr12_-_110883346 | 0.25 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr7_+_23210760 | 0.24 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr15_-_49912987 | 0.24 |
ENST00000560246.1
ENST00000558594.1 |
FAM227B
|
family with sequence similarity 227, member B |
| chr8_+_77318769 | 0.22 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr5_+_75904950 | 0.21 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr8_+_101349823 | 0.20 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr3_-_178969403 | 0.19 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr9_+_67968793 | 0.19 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr6_+_31916733 | 0.19 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr12_+_9102632 | 0.18 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr10_+_7745232 | 0.18 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7745303 | 0.17 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_+_155484155 | 0.17 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr2_-_136743436 | 0.15 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr14_-_45252031 | 0.15 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr22_-_29107919 | 0.14 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr1_+_207277590 | 0.14 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chrM_+_10464 | 0.14 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_-_143567262 | 0.14 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr14_+_21785693 | 0.13 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr2_+_191792376 | 0.13 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr3_+_101659682 | 0.13 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr14_+_61995722 | 0.13 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr2_-_191115229 | 0.13 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr11_+_29181503 | 0.12 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr9_+_6645887 | 0.12 |
ENST00000413145.1
|
RP11-390F4.6
|
RP11-390F4.6 |
| chr6_+_126102292 | 0.12 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr7_-_123197733 | 0.12 |
ENST00000470123.1
ENST00000471770.1 |
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr10_+_35484053 | 0.11 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr17_+_68047418 | 0.11 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr15_+_49913201 | 0.11 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr8_+_27629459 | 0.11 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr15_-_49913126 | 0.11 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr5_-_39270725 | 0.11 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr4_+_155484103 | 0.11 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr14_+_62164340 | 0.10 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr14_+_45605157 | 0.10 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr10_-_14596140 | 0.10 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr14_+_45605127 | 0.10 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr18_+_48918368 | 0.10 |
ENST00000583982.1
ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1
|
RP11-267C16.1 |
| chr1_-_11918988 | 0.10 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr8_+_40010989 | 0.10 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr8_+_93895865 | 0.10 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr3_-_178984759 | 0.10 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr7_+_16793160 | 0.09 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr1_-_150669500 | 0.09 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_+_132952112 | 0.09 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr14_+_35591020 | 0.09 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr2_-_40006289 | 0.09 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr5_-_58652788 | 0.09 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr4_+_57276661 | 0.09 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr15_+_100106155 | 0.09 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr12_+_20968608 | 0.08 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr11_+_108094786 | 0.08 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr1_+_207277632 | 0.08 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr14_-_35591156 | 0.08 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr1_+_171939240 | 0.08 |
ENST00000523513.1
|
DNM3
|
dynamin 3 |
| chr17_-_41277467 | 0.08 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr4_-_65275162 | 0.08 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr15_+_100106244 | 0.08 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr8_-_97247759 | 0.08 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr10_+_62538089 | 0.08 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr13_+_107028897 | 0.08 |
ENST00000439790.1
ENST00000435024.1 |
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr13_-_107214291 | 0.08 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr18_+_13612613 | 0.08 |
ENST00000586765.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_+_124824000 | 0.07 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chrM_+_10758 | 0.07 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_-_17380630 | 0.07 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr4_+_71570430 | 0.07 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_-_15548591 | 0.07 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr7_+_77469439 | 0.07 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr3_-_10334585 | 0.07 |
ENST00000430179.1
ENST00000449238.2 ENST00000437422.2 ENST00000287656.7 ENST00000457360.1 ENST00000439975.2 ENST00000446937.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr2_+_69201705 | 0.07 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr5_-_34916871 | 0.07 |
ENST00000382038.2
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr14_+_23025534 | 0.07 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr2_+_143635067 | 0.07 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr8_+_86099884 | 0.07 |
ENST00000517476.1
ENST00000521429.1 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr19_+_9296279 | 0.07 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr17_+_41322483 | 0.07 |
ENST00000341165.6
ENST00000586650.1 ENST00000422280.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr3_+_141144963 | 0.07 |
ENST00000510726.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_+_193310918 | 0.07 |
ENST00000361908.3
ENST00000392438.3 ENST00000361510.2 ENST00000361715.2 ENST00000361828.2 ENST00000361150.2 |
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr2_+_29336855 | 0.07 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr22_-_21213676 | 0.06 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr16_+_56485402 | 0.06 |
ENST00000566157.1
ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr9_+_106856831 | 0.06 |
ENST00000303219.8
ENST00000374787.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr9_-_215744 | 0.06 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr8_-_18666360 | 0.06 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_+_102615416 | 0.06 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr22_+_17956618 | 0.06 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr15_+_49913175 | 0.06 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr3_+_138340067 | 0.06 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_-_57967854 | 0.06 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr7_+_143771275 | 0.06 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr3_-_182817297 | 0.06 |
ENST00000539926.1
ENST00000476176.1 |
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr10_-_5638048 | 0.06 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr3_-_183145873 | 0.06 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr11_-_14379997 | 0.05 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr2_+_216974020 | 0.05 |
ENST00000392132.2
ENST00000417391.1 |
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr5_-_75919253 | 0.05 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr14_+_67291158 | 0.05 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chrX_+_1733876 | 0.05 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr10_+_5090940 | 0.05 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr14_-_82000140 | 0.05 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr14_+_39735411 | 0.05 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr6_-_32908765 | 0.05 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr3_+_141144954 | 0.05 |
ENST00000441582.2
ENST00000321464.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr9_-_98189055 | 0.05 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr10_-_114206649 | 0.05 |
ENST00000369404.3
ENST00000369405.3 |
ZDHHC6
|
zinc finger, DHHC-type containing 6 |
| chr17_-_39203519 | 0.05 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr13_+_108870714 | 0.05 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr3_+_169755715 | 0.05 |
ENST00000355897.5
|
GPR160
|
G protein-coupled receptor 160 |
| chr2_+_182850551 | 0.05 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr14_-_24553834 | 0.05 |
ENST00000397002.2
|
NRL
|
neural retina leucine zipper |
| chr2_-_47143160 | 0.05 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr21_+_25801041 | 0.05 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr16_-_49890016 | 0.05 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chrX_+_11777671 | 0.05 |
ENST00000380693.3
ENST00000380692.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr14_+_76618242 | 0.05 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr10_+_60028818 | 0.04 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr11_+_101785727 | 0.04 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr17_+_30348024 | 0.04 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr4_-_175204765 | 0.04 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr15_-_54025300 | 0.04 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr3_-_25915189 | 0.04 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr17_-_40170506 | 0.04 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr3_+_184080387 | 0.04 |
ENST00000455712.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr18_+_21032781 | 0.04 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr3_+_138340049 | 0.04 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_+_62860581 | 0.04 |
ENST00000393632.2
ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2
|
MON2 homolog (S. cerevisiae) |
| chr15_+_45028719 | 0.04 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chrM_+_10053 | 0.04 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrX_+_114874727 | 0.04 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr14_+_32963433 | 0.04 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr13_+_113699029 | 0.03 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr22_+_21213771 | 0.03 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chrX_+_52235228 | 0.03 |
ENST00000518075.1
|
XAGE1B
|
X antigen family, member 1B |
| chr1_+_24645865 | 0.03 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr15_+_66874502 | 0.03 |
ENST00000558797.1
|
RP11-321F6.1
|
HCG2003567; Uncharacterized protein |
| chr7_+_26332645 | 0.03 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr4_+_175204818 | 0.03 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr10_+_5005445 | 0.03 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr7_+_134331550 | 0.03 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr12_-_45270151 | 0.03 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr1_-_100643765 | 0.03 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr18_-_21891460 | 0.03 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr8_+_21777243 | 0.03 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr3_+_155860751 | 0.03 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr15_+_41245160 | 0.03 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr2_+_102927962 | 0.03 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr5_+_179125907 | 0.03 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr12_+_75874580 | 0.03 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr9_-_70490107 | 0.03 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr6_-_29395509 | 0.03 |
ENST00000377147.2
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1 |
| chr12_-_45270077 | 0.03 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr1_+_24645807 | 0.03 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_+_41323204 | 0.02 |
ENST00000542611.1
ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr11_+_114310102 | 0.02 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr1_-_89357179 | 0.02 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr15_-_34628951 | 0.02 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr12_-_9102549 | 0.02 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr2_+_201997595 | 0.02 |
ENST00000470178.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_-_113478603 | 0.02 |
ENST00000443580.1
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr3_+_193311105 | 0.02 |
ENST00000392436.2
|
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr4_-_83765613 | 0.02 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr4_+_175204865 | 0.02 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr6_+_10634158 | 0.02 |
ENST00000379591.3
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr6_-_86099898 | 0.02 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr15_-_49912944 | 0.02 |
ENST00000559905.1
|
FAM227B
|
family with sequence similarity 227, member B |
| chr4_-_48782259 | 0.02 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr5_+_147774275 | 0.02 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr2_+_201997676 | 0.02 |
ENST00000462763.1
ENST00000479953.2 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_-_136594740 | 0.02 |
ENST00000264162.2
|
LCT
|
lactase |
| chr20_-_61885826 | 0.02 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr5_+_179125368 | 0.02 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chr8_-_91095099 | 0.02 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr17_+_7308172 | 0.02 |
ENST00000575301.1
|
NLGN2
|
neuroligin 2 |
| chr17_+_53344945 | 0.02 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr12_-_14967095 | 0.02 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr5_+_136070614 | 0.02 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr8_+_117778736 | 0.02 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr4_-_47983519 | 0.02 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr4_+_130014836 | 0.02 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr15_+_100106126 | 0.02 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr10_+_18689637 | 0.01 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr11_+_57308979 | 0.01 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr1_+_156123318 | 0.01 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr6_-_53409890 | 0.01 |
ENST00000229416.6
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit |
| chr4_-_110723335 | 0.01 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr17_+_53345080 | 0.01 |
ENST00000572002.1
|
HLF
|
hepatic leukemia factor |
| chr11_+_110225855 | 0.01 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr14_+_38264418 | 0.01 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr2_+_179149636 | 0.01 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr3_+_44916098 | 0.01 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr15_+_45028753 | 0.01 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr9_+_91933407 | 0.01 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chr20_+_58571419 | 0.01 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr6_+_53964336 | 0.01 |
ENST00000447836.2
ENST00000511678.1 |
MLIP
|
muscular LMNA-interacting protein |
| chrX_-_72097698 | 0.01 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.2 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0040010 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |