Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
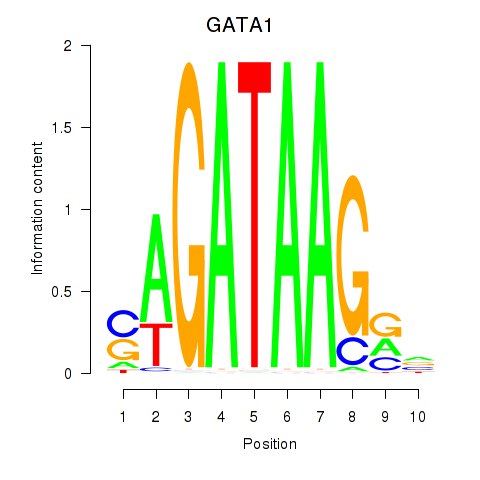
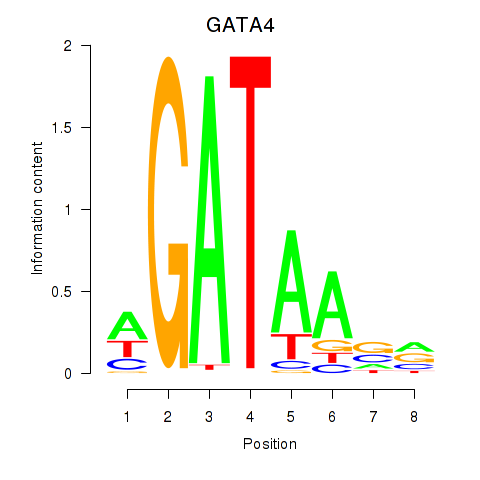
Results for GATA1_GATA4
Z-value: 0.50
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA4 | hg19_v2_chr8_+_11534462_11534475 | -0.55 | 2.6e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_202298268 | 0.37 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr1_-_235098935 | 0.31 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr1_-_235098861 | 0.26 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr2_-_14541060 | 0.25 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr13_+_24144796 | 0.25 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr19_-_13213662 | 0.24 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr10_-_112255945 | 0.22 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr2_+_189156638 | 0.22 |
ENST00000410051.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_-_135290651 | 0.21 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_88188254 | 0.21 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr8_+_27629459 | 0.21 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_+_103876528 | 0.21 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr4_-_103266219 | 0.21 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_42630389 | 0.20 |
ENST00000357001.2
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin) |
| chr21_-_43786634 | 0.20 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr10_+_118349920 | 0.19 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr3_-_135916073 | 0.19 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr2_+_192543694 | 0.19 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr10_-_96122682 | 0.19 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr17_-_71258019 | 0.18 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr8_+_107630340 | 0.17 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr11_+_34642656 | 0.17 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr2_+_3642545 | 0.17 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chr12_+_9144626 | 0.17 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr12_-_10022735 | 0.17 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr2_+_189156721 | 0.17 |
ENST00000409927.1
ENST00000409805.1 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr3_+_172468505 | 0.17 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr4_+_76871883 | 0.17 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr17_+_19314432 | 0.16 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chr12_+_56477093 | 0.16 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr11_-_10920838 | 0.16 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr19_-_55669093 | 0.16 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr2_-_44065889 | 0.16 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr12_+_4430371 | 0.15 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr13_-_49975632 | 0.15 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr11_-_10920714 | 0.15 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr9_+_33240157 | 0.15 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr1_+_199996733 | 0.15 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr15_+_49715449 | 0.14 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr2_+_114163945 | 0.14 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr14_+_61449197 | 0.14 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr20_-_60294804 | 0.14 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr7_-_99332719 | 0.14 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr5_-_82373260 | 0.14 |
ENST00000502346.1
|
TMEM167A
|
transmembrane protein 167A |
| chr11_-_125648690 | 0.14 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr11_+_64052692 | 0.14 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chrX_+_118892545 | 0.14 |
ENST00000343905.3
|
SOWAHD
|
sosondowah ankyrin repeat domain family member D |
| chr6_+_76330355 | 0.14 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr7_+_94536514 | 0.13 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_-_1036706 | 0.13 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr12_-_10324716 | 0.13 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr10_-_21186144 | 0.13 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr5_+_135496675 | 0.13 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr14_-_38036271 | 0.13 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr10_+_94594351 | 0.13 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr14_+_102276192 | 0.13 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr16_+_333152 | 0.13 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chrX_-_78622805 | 0.12 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr1_-_159880159 | 0.12 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr19_-_39303576 | 0.12 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr1_-_238108575 | 0.12 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr15_+_49913201 | 0.12 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr19_-_9879293 | 0.12 |
ENST00000397902.2
ENST00000592859.1 ENST00000588267.1 |
ZNF846
|
zinc finger protein 846 |
| chr4_-_89619386 | 0.12 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr15_+_49715293 | 0.12 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr5_-_87516448 | 0.12 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr5_+_32788945 | 0.11 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chrX_-_84634737 | 0.11 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr21_-_43816152 | 0.11 |
ENST00000433957.2
ENST00000398397.3 |
TMPRSS3
|
transmembrane protease, serine 3 |
| chr8_-_30002179 | 0.11 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr5_+_40841410 | 0.11 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chrX_+_38420623 | 0.11 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr13_+_24144509 | 0.10 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_+_102276209 | 0.10 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_+_74379083 | 0.10 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr3_+_172468749 | 0.10 |
ENST00000366254.2
ENST00000415665.1 ENST00000438041.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr14_-_45603657 | 0.10 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr16_+_28857916 | 0.10 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr19_+_37808831 | 0.10 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr5_-_157161727 | 0.10 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr10_-_71993176 | 0.10 |
ENST00000373232.3
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr12_+_6419877 | 0.10 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr3_-_10334585 | 0.10 |
ENST00000430179.1
ENST00000449238.2 ENST00000437422.2 ENST00000287656.7 ENST00000457360.1 ENST00000439975.2 ENST00000446937.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr2_-_219850277 | 0.10 |
ENST00000295727.1
|
FEV
|
FEV (ETS oncogene family) |
| chr3_-_194188956 | 0.10 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr12_-_54689532 | 0.09 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr11_+_10326918 | 0.09 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr2_+_170440844 | 0.09 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr1_-_167487758 | 0.09 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr1_+_76540386 | 0.09 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr6_-_30128657 | 0.09 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr11_+_76571911 | 0.09 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr19_-_11494975 | 0.09 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr12_-_21928515 | 0.09 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr6_-_56112235 | 0.09 |
ENST00000370817.3
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr4_+_74606223 | 0.09 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr12_+_90313394 | 0.09 |
ENST00000549551.1
|
RP11-654D12.2
|
RP11-654D12.2 |
| chr2_+_189156586 | 0.08 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr1_-_27240455 | 0.08 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr9_-_20382446 | 0.08 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_+_220379052 | 0.08 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr12_+_109569155 | 0.08 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr5_-_141703713 | 0.08 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr1_-_167487808 | 0.08 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr15_+_71184931 | 0.08 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chrX_+_100224676 | 0.08 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr3_-_10334617 | 0.08 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr1_+_199996702 | 0.08 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr4_-_83483360 | 0.08 |
ENST00000449862.2
|
TMEM150C
|
transmembrane protein 150C |
| chr12_+_104359576 | 0.08 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr4_-_100815525 | 0.08 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr5_+_67576062 | 0.08 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_+_7533439 | 0.08 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr2_-_74555350 | 0.08 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr16_+_28857957 | 0.08 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_39339777 | 0.08 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr9_-_116159635 | 0.08 |
ENST00000452726.1
|
ALAD
|
aminolevulinate dehydratase |
| chr4_-_103266355 | 0.08 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_109849612 | 0.08 |
ENST00000357155.1
|
MYBPHL
|
myosin binding protein H-like |
| chr4_-_144826682 | 0.07 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr17_-_62084241 | 0.07 |
ENST00000449662.2
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr10_+_28822236 | 0.07 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr21_-_35014027 | 0.07 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr7_+_12727250 | 0.07 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr11_-_105892937 | 0.07 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr3_-_33138592 | 0.07 |
ENST00000415454.1
|
GLB1
|
galactosidase, beta 1 |
| chr6_-_31514516 | 0.07 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr3_+_148447887 | 0.07 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr4_-_76439483 | 0.07 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_+_145780739 | 0.07 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_+_158690089 | 0.07 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chrM_+_14741 | 0.07 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr14_-_23285069 | 0.07 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr10_-_104597286 | 0.07 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr8_+_32579321 | 0.07 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr12_-_104359475 | 0.07 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr2_+_160590469 | 0.07 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr12_-_91546926 | 0.07 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_-_57233966 | 0.07 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr11_-_85393886 | 0.07 |
ENST00000534224.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr15_-_99057551 | 0.07 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr6_-_10115007 | 0.07 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr7_+_150549565 | 0.07 |
ENST00000360937.4
ENST00000416793.2 ENST00000483043.1 |
AOC1
|
amine oxidase, copper containing 1 |
| chr16_-_66907139 | 0.07 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr19_-_4717835 | 0.07 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chrX_+_84258832 | 0.07 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr1_-_11918988 | 0.07 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr17_+_38673270 | 0.07 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr2_+_187350883 | 0.06 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr16_-_46782221 | 0.06 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr8_+_76452097 | 0.06 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_209859510 | 0.06 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr7_+_120629653 | 0.06 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_-_67981870 | 0.06 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr7_-_1980128 | 0.06 |
ENST00000437877.1
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr7_-_27169801 | 0.06 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr14_-_85996332 | 0.06 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr1_-_100643765 | 0.06 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chrX_-_63615297 | 0.06 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr19_-_41934635 | 0.06 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr2_+_33701286 | 0.06 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_225615599 | 0.06 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr12_+_121088291 | 0.06 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
| chr5_+_139505520 | 0.06 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr7_-_143892748 | 0.06 |
ENST00000378115.2
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr10_-_129691195 | 0.06 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr14_-_53258180 | 0.06 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_-_140179276 | 0.06 |
ENST00000443720.2
ENST00000255977.2 |
MKRN1
|
makorin ring finger protein 1 |
| chr5_-_20575959 | 0.06 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr19_-_13213954 | 0.06 |
ENST00000590974.1
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr15_-_77197781 | 0.06 |
ENST00000564590.1
|
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr18_+_61575200 | 0.06 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr8_+_42873548 | 0.06 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr14_+_53173890 | 0.06 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr7_+_117864708 | 0.06 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr13_-_31191642 | 0.06 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr5_+_50679506 | 0.06 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr1_+_145516252 | 0.06 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr5_+_68513622 | 0.06 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr16_+_14280564 | 0.06 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr5_+_67584523 | 0.06 |
ENST00000521409.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_+_42665742 | 0.06 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr13_+_53029564 | 0.06 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr12_-_121476750 | 0.06 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr17_-_47841485 | 0.06 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr6_-_150039249 | 0.06 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr3_-_24207039 | 0.06 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr3_-_56717246 | 0.05 |
ENST00000355628.5
|
FAM208A
|
family with sequence similarity 208, member A |
| chr2_-_197041193 | 0.05 |
ENST00000409228.1
|
STK17B
|
serine/threonine kinase 17b |
| chr2_+_170440902 | 0.05 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr15_-_64665911 | 0.05 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr10_-_63995871 | 0.05 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr16_+_4838412 | 0.05 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr17_-_60142609 | 0.05 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr12_-_123634449 | 0.05 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr5_+_56471592 | 0.05 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr13_-_96329048 | 0.05 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr4_-_83821676 | 0.05 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chrX_-_49121165 | 0.05 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr15_-_52861394 | 0.05 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.2 | GO:1990768 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.0 | 0.0 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0044027 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0002669 | regulation of tolerance induction dependent upon immune response(GO:0002652) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |