Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
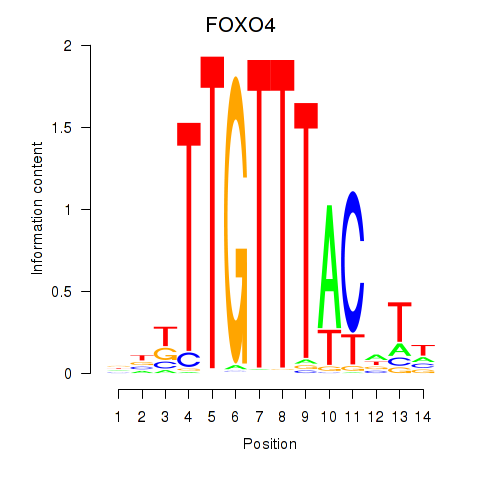
Results for FOXO4
Z-value: 0.79
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | -0.45 | 3.7e-01 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_134638767 | 0.56 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_-_39150385 | 0.47 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr4_-_85771168 | 0.45 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr3_+_187461442 | 0.45 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr1_+_95616933 | 0.45 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr16_+_28857916 | 0.43 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr9_-_15472730 | 0.39 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr16_-_79804435 | 0.38 |
ENST00000562921.1
ENST00000566729.1 |
RP11-345M22.2
|
RP11-345M22.2 |
| chr15_+_63188009 | 0.36 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr14_+_102276192 | 0.35 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_+_24661479 | 0.34 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr4_+_76871883 | 0.32 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr2_-_197458323 | 0.31 |
ENST00000452031.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr12_+_28605426 | 0.27 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr5_-_68339648 | 0.26 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr6_+_8652370 | 0.25 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr18_-_53303123 | 0.24 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr8_+_101349823 | 0.23 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr4_-_159080806 | 0.23 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr12_-_100656134 | 0.23 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr2_+_58134756 | 0.22 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr6_-_152623231 | 0.22 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr5_+_40841410 | 0.21 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr15_-_52970820 | 0.21 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr4_+_78804393 | 0.21 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr2_+_196521845 | 0.20 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_-_44828838 | 0.20 |
ENST00000560750.1
|
EIF3J-AS1
|
EIF3J antisense RNA 1 (head to head) |
| chr2_+_42104692 | 0.20 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr9_-_86432547 | 0.19 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr9_+_42671887 | 0.19 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr7_+_7811992 | 0.19 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr6_+_143447322 | 0.19 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr1_+_43855545 | 0.19 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr7_+_37723336 | 0.19 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr10_-_13570533 | 0.19 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr4_+_95174445 | 0.19 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_68696652 | 0.18 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr11_+_33061336 | 0.18 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr7_-_95064264 | 0.17 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr5_+_60933634 | 0.17 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr5_-_137674000 | 0.17 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr8_+_74903580 | 0.17 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr8_+_31497271 | 0.17 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr16_+_28857957 | 0.17 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr6_+_62284008 | 0.16 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chrX_-_148676974 | 0.16 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr2_+_152266392 | 0.16 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr5_+_133706865 | 0.16 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr8_-_30002179 | 0.16 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr10_-_25241499 | 0.16 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr14_-_92572894 | 0.16 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr7_-_38394118 | 0.16 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chrX_+_106045891 | 0.16 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr13_+_24144796 | 0.16 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr3_-_57530051 | 0.16 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr18_+_56806701 | 0.15 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr19_+_13134772 | 0.15 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr8_-_123793048 | 0.15 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr6_-_88038996 | 0.15 |
ENST00000525899.1
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chrX_+_148855726 | 0.15 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr1_-_234667504 | 0.15 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr2_+_175260514 | 0.15 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr11_-_10879572 | 0.15 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr5_-_42811986 | 0.14 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr10_-_61900762 | 0.14 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr6_-_117747015 | 0.14 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr6_+_134273300 | 0.14 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr11_-_85780853 | 0.14 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr13_+_98086445 | 0.14 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr17_+_7533439 | 0.14 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr4_-_164534657 | 0.14 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr3_-_71777824 | 0.14 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr2_+_182850551 | 0.14 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr11_+_77532233 | 0.14 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr7_-_95225768 | 0.14 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr14_+_102276209 | 0.13 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr5_+_79703823 | 0.13 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr18_-_53735601 | 0.13 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr3_+_108015382 | 0.13 |
ENST00000463019.3
ENST00000491820.1 ENST00000467562.1 ENST00000482430.1 ENST00000462629.1 |
HHLA2
|
HERV-H LTR-associating 2 |
| chr8_+_125860939 | 0.13 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chrX_+_100743031 | 0.13 |
ENST00000423738.3
|
ARMCX4
|
armadillo repeat containing, X-linked 4 |
| chr11_-_10879593 | 0.13 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr4_-_21950356 | 0.13 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr3_+_148447887 | 0.12 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_153940741 | 0.12 |
ENST00000431292.1
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr8_+_93895865 | 0.12 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr11_-_46141338 | 0.12 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr10_-_5638048 | 0.11 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr18_-_68004529 | 0.11 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr8_-_6420759 | 0.11 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr1_+_145525015 | 0.11 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr13_-_40924439 | 0.11 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr7_-_14028488 | 0.11 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr10_+_111985713 | 0.11 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr8_-_133772794 | 0.11 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr17_+_8924837 | 0.11 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr7_+_37723450 | 0.10 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr12_-_100486668 | 0.10 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr11_+_46193466 | 0.10 |
ENST00000533793.1
|
RP11-702F3.3
|
RP11-702F3.3 |
| chr7_+_99425633 | 0.10 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr12_-_31479045 | 0.10 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr3_-_28390120 | 0.10 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr10_+_70847852 | 0.10 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr2_-_148779106 | 0.10 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr10_-_4285923 | 0.10 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr3_+_169490606 | 0.10 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr13_-_41240717 | 0.10 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr3_-_101232019 | 0.10 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr4_-_140223670 | 0.10 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_-_102591604 | 0.10 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr2_+_12857015 | 0.10 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr20_-_1309809 | 0.10 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr5_+_65222500 | 0.10 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr2_-_71454185 | 0.09 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr6_+_126277842 | 0.09 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr12_+_96588279 | 0.09 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr6_+_21666633 | 0.09 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr2_-_113012592 | 0.09 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
| chr9_+_67977438 | 0.09 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr17_+_61151306 | 0.09 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_+_101292939 | 0.09 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr1_+_97188188 | 0.09 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr14_-_64108125 | 0.09 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr1_+_6845384 | 0.09 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr1_-_150669500 | 0.09 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr3_+_112930946 | 0.09 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr3_-_28390581 | 0.08 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr14_+_96968707 | 0.08 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr18_+_47088401 | 0.08 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr2_+_12857043 | 0.08 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr6_-_127780510 | 0.08 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr4_-_70518941 | 0.08 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr7_+_106505912 | 0.08 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr7_+_129906660 | 0.08 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr4_-_111120334 | 0.08 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr1_+_236686717 | 0.08 |
ENST00000341872.6
ENST00000450372.2 |
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr14_-_55878538 | 0.08 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr4_-_147442982 | 0.08 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr7_+_77469439 | 0.08 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr17_-_40134339 | 0.08 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr5_+_137673945 | 0.08 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr2_+_149402989 | 0.08 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrX_-_122756660 | 0.08 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr2_+_149402553 | 0.07 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr3_-_28390298 | 0.07 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_145940126 | 0.07 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr11_-_119247004 | 0.07 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr5_+_64920543 | 0.07 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr15_+_49462434 | 0.07 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr22_-_45809495 | 0.07 |
ENST00000404354.3
|
SMC1B
|
structural maintenance of chromosomes 1B |
| chr17_-_49021974 | 0.07 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr13_-_31038370 | 0.07 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr9_-_88897426 | 0.07 |
ENST00000375991.4
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr19_-_36523529 | 0.07 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr17_-_8263538 | 0.07 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr16_-_65106110 | 0.07 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr15_-_31283798 | 0.07 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr6_+_32121908 | 0.07 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_85397167 | 0.07 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr5_+_135468516 | 0.07 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr13_+_24144509 | 0.07 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr5_-_133706695 | 0.07 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr14_+_102276132 | 0.07 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr2_+_66662510 | 0.07 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr15_-_52971544 | 0.07 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr3_+_99536663 | 0.07 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr22_-_45809446 | 0.07 |
ENST00000357450.4
|
SMC1B
|
structural maintenance of chromosomes 1B |
| chr2_-_25564750 | 0.07 |
ENST00000321117.5
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr8_-_72459885 | 0.07 |
ENST00000523987.1
|
RP11-1102P16.1
|
Uncharacterized protein |
| chr12_+_96588143 | 0.06 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr14_+_96968802 | 0.06 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr1_+_174670143 | 0.06 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr15_+_43885252 | 0.06 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr6_+_47666275 | 0.06 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr14_-_61124977 | 0.06 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr1_+_10490127 | 0.06 |
ENST00000602787.1
ENST00000602296.1 ENST00000400900.2 |
APITD1
APITD1-CORT
|
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr2_+_162165038 | 0.06 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr17_-_30185971 | 0.06 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr7_+_73242490 | 0.06 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr6_-_105627735 | 0.06 |
ENST00000254765.3
|
POPDC3
|
popeye domain containing 3 |
| chr1_-_39395165 | 0.06 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr1_-_31661000 | 0.06 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr2_+_65283506 | 0.06 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr2_+_58655461 | 0.06 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr4_-_147442817 | 0.06 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr10_-_37891859 | 0.06 |
ENST00000544824.1
|
MTRNR2L7
|
MT-RNR2-like 7 |
| chr14_-_102605983 | 0.06 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr3_-_64009658 | 0.06 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr20_+_44035847 | 0.06 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chrX_-_70329118 | 0.06 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr2_-_225434538 | 0.06 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr12_-_7596735 | 0.06 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr9_-_95186739 | 0.06 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr1_-_53608249 | 0.06 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr3_-_28390415 | 0.06 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr19_-_14196574 | 0.06 |
ENST00000548523.1
ENST00000343945.5 |
C19orf67
|
chromosome 19 open reading frame 67 |
| chr14_-_99737822 | 0.06 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_+_160765919 | 0.06 |
ENST00000341032.4
ENST00000368041.2 ENST00000368040.1 |
LY9
|
lymphocyte antigen 9 |
| chr10_-_82049424 | 0.06 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr1_+_150229554 | 0.06 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.1 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.0 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) hindlimb morphogenesis(GO:0035137) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |