Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
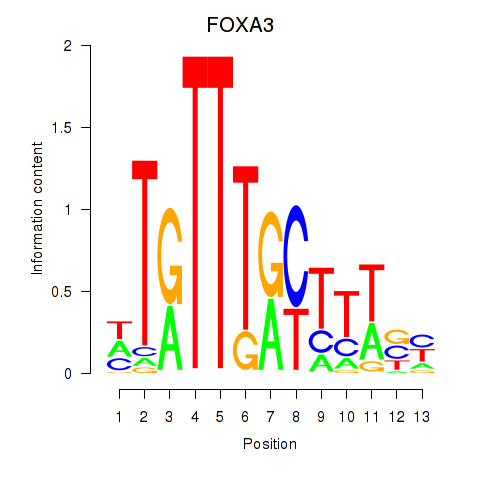
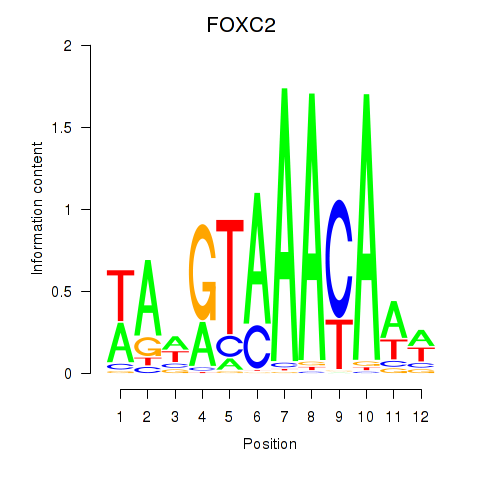
Results for FOXA3_FOXC2
Z-value: 0.74
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA3 | hg19_v2_chr19_+_46367518_46367567 | 0.14 | 7.9e-01 | Click! |
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | 0.10 | 8.5e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_62186498 | 0.47 |
ENST00000278282.2
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr8_+_97597148 | 0.36 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr17_-_26695013 | 0.34 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr12_-_57328187 | 0.34 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr8_+_101349823 | 0.33 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr3_-_193096600 | 0.31 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr17_-_77967433 | 0.31 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr12_+_28605426 | 0.27 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr15_+_38964048 | 0.26 |
ENST00000560203.1
ENST00000557946.1 |
RP11-275I4.2
|
RP11-275I4.2 |
| chr2_+_58655461 | 0.25 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr2_-_87248975 | 0.22 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr1_+_152486950 | 0.21 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr5_+_60933634 | 0.20 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr8_-_30002179 | 0.20 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr12_+_59989918 | 0.20 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_+_142829162 | 0.19 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr18_+_61575200 | 0.18 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr1_+_174933899 | 0.17 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_+_96588279 | 0.17 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr18_+_68002675 | 0.16 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr1_+_239882842 | 0.16 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr2_-_175711924 | 0.16 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr16_-_67597789 | 0.16 |
ENST00000605277.1
|
CTD-2012K14.6
|
CTD-2012K14.6 |
| chr17_-_39150385 | 0.16 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr5_+_133859996 | 0.16 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr5_+_40841276 | 0.15 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr10_+_124739911 | 0.15 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr5_-_137674000 | 0.15 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr14_-_94789663 | 0.15 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr7_+_151791074 | 0.15 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_+_20650796 | 0.15 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chr4_+_76871883 | 0.15 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr13_+_95364963 | 0.15 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr3_-_187455680 | 0.14 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr9_+_116343192 | 0.14 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr3_-_71114066 | 0.14 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr4_-_164534657 | 0.13 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr12_-_12714006 | 0.13 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr15_+_69857515 | 0.13 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr5_+_125706998 | 0.13 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr15_+_69854027 | 0.13 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr1_+_104159999 | 0.11 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr13_+_50589390 | 0.11 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr1_+_101361782 | 0.11 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr7_+_129906660 | 0.10 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr7_+_95115210 | 0.10 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_+_151791037 | 0.10 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr18_+_59415396 | 0.10 |
ENST00000567801.1
|
RP11-1096D5.1
|
RP11-1096D5.1 |
| chr7_+_134832808 | 0.10 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr3_-_105588231 | 0.10 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_+_124571409 | 0.09 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr4_-_153303658 | 0.09 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr17_+_72426891 | 0.09 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr21_+_17791648 | 0.09 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_71551652 | 0.09 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr4_+_57138437 | 0.09 |
ENST00000504228.1
ENST00000541073.1 |
KIAA1211
|
KIAA1211 |
| chr21_+_17791838 | 0.09 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_+_75335604 | 0.09 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr7_+_28452130 | 0.09 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr9_+_131062367 | 0.09 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr2_+_220042933 | 0.09 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr12_-_53207842 | 0.09 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr4_+_165675269 | 0.09 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr1_-_149859466 | 0.09 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr17_-_26694979 | 0.09 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr6_-_41715128 | 0.08 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr2_+_175260514 | 0.08 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr19_-_36019123 | 0.08 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr4_-_87028478 | 0.08 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_-_140223670 | 0.08 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chrX_-_15402498 | 0.08 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr14_+_20215587 | 0.08 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr9_-_20382446 | 0.08 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr18_+_47088401 | 0.08 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr4_-_74486109 | 0.08 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_-_120354079 | 0.08 |
ENST00000354219.1
ENST00000369401.4 ENST00000256585.5 |
REG4
|
regenerating islet-derived family, member 4 |
| chr9_-_95186739 | 0.08 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr8_+_52730143 | 0.08 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr10_+_105314881 | 0.08 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr5_+_95998714 | 0.08 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr11_-_116708302 | 0.07 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr1_+_154540246 | 0.07 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr1_+_12806141 | 0.07 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chrX_+_24167746 | 0.07 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr10_-_75168071 | 0.07 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr1_-_146696901 | 0.07 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr17_+_8924837 | 0.07 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr4_-_186456652 | 0.07 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr2_-_21266816 | 0.07 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr1_-_179851611 | 0.07 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr10_-_63995871 | 0.07 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr2_-_208030295 | 0.07 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_-_108464321 | 0.07 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr10_-_4285923 | 0.07 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr20_+_9049742 | 0.07 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr12_+_27849378 | 0.07 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr12_-_76879852 | 0.07 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr12_+_58087738 | 0.07 |
ENST00000552285.1
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr4_-_140223614 | 0.07 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr18_-_25616519 | 0.07 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr3_+_69985792 | 0.07 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_172290482 | 0.06 |
ENST00000442541.1
ENST00000392599.2 ENST00000375258.4 |
METTL8
|
methyltransferase like 8 |
| chr12_-_21928515 | 0.06 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr1_+_172389821 | 0.06 |
ENST00000367727.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr20_+_11898507 | 0.06 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr2_-_46844242 | 0.06 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr1_+_40974431 | 0.06 |
ENST00000296380.4
ENST00000432259.1 ENST00000418186.1 |
EXO5
|
exonuclease 5 |
| chr7_-_111424462 | 0.06 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_+_71587669 | 0.06 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr5_+_61708488 | 0.06 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr2_-_145275828 | 0.06 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_95385819 | 0.06 |
ENST00000507997.1
|
RP11-254I22.1
|
RP11-254I22.1 |
| chr11_-_107582775 | 0.06 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr1_-_223308098 | 0.06 |
ENST00000342210.6
|
TLR5
|
toll-like receptor 5 |
| chr4_-_88450372 | 0.06 |
ENST00000543631.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr6_-_135818368 | 0.06 |
ENST00000367798.2
|
AHI1
|
Abelson helper integration site 1 |
| chr7_+_73242490 | 0.06 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr1_+_43855545 | 0.06 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr15_+_58430368 | 0.06 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chrX_+_107288239 | 0.06 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_+_79567233 | 0.06 |
ENST00000514130.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr5_+_137673945 | 0.06 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr11_-_46615498 | 0.06 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr1_-_149459549 | 0.06 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr2_-_175711978 | 0.06 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr2_+_28974531 | 0.06 |
ENST00000420282.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_-_71532668 | 0.06 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr2_+_128175997 | 0.06 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chrX_+_107288197 | 0.06 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_-_22259845 | 0.06 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr8_+_104384616 | 0.06 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr3_+_132316081 | 0.05 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr20_-_55841662 | 0.05 |
ENST00000395863.3
ENST00000450594.2 |
BMP7
|
bone morphogenetic protein 7 |
| chr3_+_119316721 | 0.05 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr12_-_21927736 | 0.05 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr2_-_145275228 | 0.05 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_39140549 | 0.05 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr3_-_71294304 | 0.05 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr4_+_110769258 | 0.05 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr15_+_59910132 | 0.05 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_-_152332480 | 0.05 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr17_-_29641104 | 0.05 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_159141397 | 0.05 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr11_-_85397167 | 0.05 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr14_-_99737822 | 0.05 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr15_+_59665194 | 0.05 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr3_+_69985734 | 0.05 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr10_-_71176623 | 0.05 |
ENST00000373306.4
|
TACR2
|
tachykinin receptor 2 |
| chr6_-_88875628 | 0.05 |
ENST00000551417.1
|
CNR1
|
cannabinoid receptor 1 (brain) |
| chr17_+_7487146 | 0.05 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr6_+_10528560 | 0.04 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr10_-_31288398 | 0.04 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr4_-_186456766 | 0.04 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr14_+_23299088 | 0.04 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr2_-_175260368 | 0.04 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr1_+_104293028 | 0.04 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr4_-_174256276 | 0.04 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr8_-_133772870 | 0.04 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr2_+_145425534 | 0.04 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_+_135468516 | 0.04 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr16_-_48281305 | 0.04 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr6_-_26056695 | 0.04 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr12_+_72080253 | 0.04 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr16_+_77233294 | 0.04 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr16_+_84801852 | 0.04 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr10_+_80008505 | 0.04 |
ENST00000434974.1
ENST00000423770.1 ENST00000432742.1 |
LINC00856
|
long intergenic non-protein coding RNA 856 |
| chr17_-_66951382 | 0.04 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_+_59664884 | 0.04 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr2_-_220042825 | 0.04 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr4_+_172734548 | 0.04 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr11_+_20044096 | 0.04 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr2_+_196521845 | 0.04 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_-_28670283 | 0.04 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr11_-_57158109 | 0.04 |
ENST00000525955.1
ENST00000533605.1 ENST00000311862.5 |
PRG2
|
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
| chrX_-_55020511 | 0.04 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr21_-_32931290 | 0.04 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr5_+_101569696 | 0.04 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr14_+_56584414 | 0.04 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr1_+_207277590 | 0.04 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr9_+_103340354 | 0.04 |
ENST00000307584.5
|
MURC
|
muscle-related coiled-coil protein |
| chr8_+_19171128 | 0.04 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr4_+_148653206 | 0.04 |
ENST00000336498.3
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr13_-_40924439 | 0.04 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr12_-_92539614 | 0.04 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr12_-_49581152 | 0.04 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_119316689 | 0.04 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr10_-_50603497 | 0.04 |
ENST00000374139.2
|
DRGX
|
dorsal root ganglia homeobox |
| chr12_-_122018346 | 0.04 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr12_-_102591604 | 0.04 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr13_-_36429763 | 0.04 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chrX_+_135730373 | 0.04 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr4_+_15341442 | 0.03 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr7_+_106505912 | 0.03 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_+_43855560 | 0.03 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr14_+_23654525 | 0.03 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chrX_+_135730297 | 0.03 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr3_+_159570722 | 0.03 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr3_+_135741576 | 0.03 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr17_-_66951474 | 0.03 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr11_-_18062335 | 0.03 |
ENST00000341556.2
|
TPH1
|
tryptophan hydroxylase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010193 | response to ozone(GO:0010193) negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0039008 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.0 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.0 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0015254 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |