Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
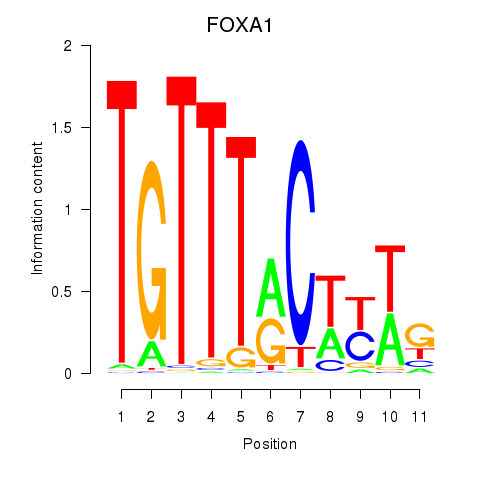
Results for FOXA1
Z-value: 0.79
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.4 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg19_v2_chr14_-_38064198_38064239 | 0.42 | 4.1e-01 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_162101247 | 0.92 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr4_-_103940791 | 0.65 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr4_-_155533787 | 0.62 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr22_-_40929812 | 0.54 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr9_-_4859260 | 0.49 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr10_+_91092241 | 0.48 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr3_-_148939598 | 0.47 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr4_+_55095428 | 0.46 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_+_116654376 | 0.45 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr3_-_148939835 | 0.44 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_-_89746264 | 0.41 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr13_-_86373536 | 0.40 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr12_-_12714025 | 0.40 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr5_+_40841410 | 0.39 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr8_-_131028869 | 0.38 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr17_-_46035187 | 0.36 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr14_+_64970662 | 0.35 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_-_1356719 | 0.34 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr12_-_92536433 | 0.33 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr3_-_145878954 | 0.33 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr1_-_1356628 | 0.32 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr17_-_26694979 | 0.31 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr5_+_150157860 | 0.31 |
ENST00000600109.1
|
AC010441.1
|
AC010441.1 |
| chr14_+_38065052 | 0.31 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr14_+_56127960 | 0.31 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_+_56127989 | 0.30 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_+_32531893 | 0.30 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr8_+_126442563 | 0.29 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr5_+_150157444 | 0.29 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr2_-_165424973 | 0.26 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr5_+_169011033 | 0.26 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr1_+_35225339 | 0.26 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr12_-_89746173 | 0.25 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr4_+_165675197 | 0.25 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr14_+_64971292 | 0.25 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr5_+_102201430 | 0.24 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_25706368 | 0.24 |
ENST00000424225.1
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa |
| chr3_+_142315225 | 0.23 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr5_+_102201509 | 0.23 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_+_26251835 | 0.23 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr11_+_65851443 | 0.23 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr8_-_74495065 | 0.23 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr16_+_21608525 | 0.22 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr10_-_32217717 | 0.22 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr5_-_41510725 | 0.22 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr11_+_1151573 | 0.22 |
ENST00000534821.1
ENST00000356191.2 |
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming |
| chr1_-_111743285 | 0.21 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr3_-_45957088 | 0.21 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr21_-_38445011 | 0.21 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_+_172950227 | 0.21 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr11_-_119247004 | 0.21 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr2_-_183291741 | 0.20 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_+_95908041 | 0.20 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr18_+_70536215 | 0.20 |
ENST00000578967.1
|
RP11-676J15.1
|
RP11-676J15.1 |
| chr17_-_57229155 | 0.20 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_+_70026795 | 0.19 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr3_+_142315294 | 0.19 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr9_-_3525968 | 0.19 |
ENST00000382004.3
ENST00000302303.1 ENST00000449190.1 |
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr13_-_31038370 | 0.18 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr1_+_199996702 | 0.18 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_-_112579869 | 0.18 |
ENST00000297145.4
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr14_+_64971438 | 0.18 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_+_57308979 | 0.18 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr5_-_133510456 | 0.18 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr1_-_115301235 | 0.18 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr2_-_166810261 | 0.17 |
ENST00000243344.7
|
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr3_+_148447887 | 0.17 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr7_+_106415457 | 0.17 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr2_+_238877424 | 0.17 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr3_+_186692745 | 0.17 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr13_+_50589390 | 0.17 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chrX_-_47863348 | 0.17 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr5_+_179921344 | 0.17 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr15_-_55541227 | 0.17 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_160394940 | 0.16 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chrX_-_20237059 | 0.16 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr1_+_35734562 | 0.16 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr19_-_56047661 | 0.16 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr2_+_47630108 | 0.15 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr1_-_11107280 | 0.15 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr2_+_58655520 | 0.15 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr18_-_52626622 | 0.15 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr16_-_87970122 | 0.15 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr10_-_81708854 | 0.15 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr1_+_40810516 | 0.15 |
ENST00000435168.2
|
SMAP2
|
small ArfGAP2 |
| chr8_+_119294456 | 0.15 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr16_+_4666475 | 0.15 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr2_+_196521845 | 0.15 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr21_+_17443521 | 0.15 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_-_127674883 | 0.14 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr3_-_129407535 | 0.14 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr8_-_131028782 | 0.14 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr6_+_101847105 | 0.14 |
ENST00000369137.3
ENST00000318991.6 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr2_+_47630255 | 0.14 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr10_-_73848086 | 0.14 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr4_-_70080449 | 0.14 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr12_+_27849378 | 0.14 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr12_+_10658489 | 0.14 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr4_+_26321284 | 0.14 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_-_772901 | 0.14 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr14_-_31926701 | 0.13 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr2_+_196521903 | 0.13 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_-_20236970 | 0.13 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr9_-_20622478 | 0.13 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr6_-_90024967 | 0.13 |
ENST00000602399.1
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr2_-_192711968 | 0.13 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr9_+_82186872 | 0.13 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr3_+_189349162 | 0.12 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr1_-_177939041 | 0.12 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr6_-_165989936 | 0.12 |
ENST00000354448.4
|
PDE10A
|
phosphodiesterase 10A |
| chr4_-_111119804 | 0.12 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr1_+_199996733 | 0.12 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr4_+_56719782 | 0.12 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr10_+_51549498 | 0.11 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr1_+_24646002 | 0.11 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr6_+_47666275 | 0.11 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr14_-_64971288 | 0.11 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr13_+_78109804 | 0.11 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr5_+_138209688 | 0.11 |
ENST00000518381.1
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr3_+_108321623 | 0.11 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr10_+_115469134 | 0.11 |
ENST00000452490.2
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr10_+_124739964 | 0.11 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr5_-_41510656 | 0.11 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_+_61547405 | 0.11 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr2_+_152266604 | 0.11 |
ENST00000430328.2
|
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr7_-_107968921 | 0.11 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_+_24645865 | 0.10 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_+_179921430 | 0.10 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr4_-_174256276 | 0.10 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr2_-_172017393 | 0.10 |
ENST00000442919.2
|
TLK1
|
tousled-like kinase 1 |
| chr11_-_116663127 | 0.10 |
ENST00000433069.1
ENST00000542499.1 |
APOA5
|
apolipoprotein A-V |
| chr7_+_116165754 | 0.10 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr6_-_31550192 | 0.10 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr9_+_126131131 | 0.10 |
ENST00000373629.2
|
CRB2
|
crumbs homolog 2 (Drosophila) |
| chr15_+_96869165 | 0.10 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_90025011 | 0.10 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr7_+_77469439 | 0.09 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr19_+_16178317 | 0.09 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr3_+_141106643 | 0.09 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_-_112579673 | 0.09 |
ENST00000432572.1
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr4_+_187148556 | 0.09 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr17_-_40897043 | 0.09 |
ENST00000428826.2
ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1
|
enhancer of zeste homolog 1 (Drosophila) |
| chr4_+_170581213 | 0.09 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr21_+_17442799 | 0.09 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_30199516 | 0.09 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr21_+_33671264 | 0.09 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr5_-_142065612 | 0.09 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr5_+_95998746 | 0.09 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr1_-_27240455 | 0.09 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr3_-_45957534 | 0.09 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr2_+_105050794 | 0.09 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr14_-_31926623 | 0.08 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr4_+_86396265 | 0.08 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_-_182545603 | 0.08 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr7_-_84121858 | 0.08 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_24645807 | 0.08 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_+_95998673 | 0.08 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr14_-_38064198 | 0.08 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr20_-_55841398 | 0.08 |
ENST00000395864.3
|
BMP7
|
bone morphogenetic protein 7 |
| chr11_-_102401469 | 0.08 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr2_-_172017343 | 0.08 |
ENST00000431350.2
ENST00000360843.3 |
TLK1
|
tousled-like kinase 1 |
| chr13_+_78109884 | 0.08 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr11_+_107799118 | 0.08 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr9_+_6413317 | 0.08 |
ENST00000276893.5
ENST00000381373.3 |
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr2_+_152266392 | 0.08 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr1_+_246729724 | 0.08 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr6_+_7107999 | 0.08 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr12_-_14849470 | 0.07 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr1_+_204839959 | 0.07 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr22_+_21128167 | 0.07 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr2_-_69180083 | 0.07 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr15_+_38964048 | 0.07 |
ENST00000560203.1
ENST00000557946.1 |
RP11-275I4.2
|
RP11-275I4.2 |
| chr7_+_134576317 | 0.07 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr20_-_45984401 | 0.07 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr8_-_133772794 | 0.07 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chrX_-_24665208 | 0.07 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr2_-_158345462 | 0.07 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr13_+_76334795 | 0.07 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr2_-_158345341 | 0.06 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr2_+_179345173 | 0.06 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chrX_+_24167828 | 0.06 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr7_+_134576151 | 0.06 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr5_+_96271141 | 0.06 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr3_+_72201910 | 0.06 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr17_+_9728828 | 0.06 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr1_+_101361782 | 0.06 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr16_-_67517716 | 0.06 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr4_+_55095264 | 0.06 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr13_-_49975632 | 0.06 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr12_+_96588143 | 0.06 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr13_+_76334567 | 0.06 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr1_+_145883868 | 0.06 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr12_+_96588368 | 0.05 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr14_-_70263979 | 0.05 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chrX_+_24167746 | 0.05 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr7_+_95115210 | 0.05 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr11_+_31833939 | 0.05 |
ENST00000530348.1
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr6_+_74405501 | 0.05 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr14_+_56585048 | 0.05 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_-_68034470 | 0.05 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.3 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.5 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.5 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.2 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.3 | GO:0010520 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.3 | GO:0045659 | eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.6 | GO:0031639 | plasminogen activation(GO:0031639) negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:1901295 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.5 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.7 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |