Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for FOSL1
Z-value: 1.17
Transcription factors associated with FOSL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL1
|
ENSG00000175592.4 | FOS like 1, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL1 | hg19_v2_chr11_-_65667997_65668044 | 0.97 | 1.3e-03 | Click! |
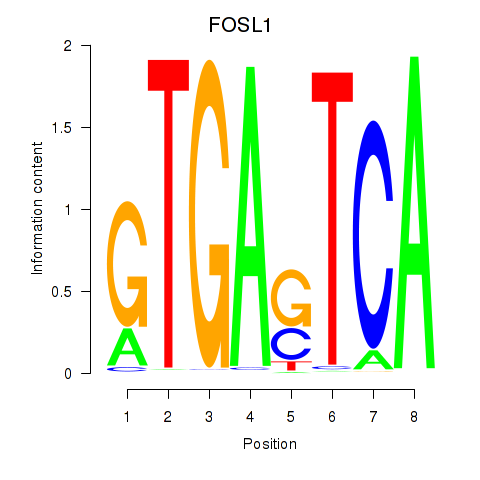
Activity profile of FOSL1 motif
Sorted Z-values of FOSL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_130597935 | 1.45 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr17_-_39769005 | 0.97 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr15_+_41062159 | 0.93 |
ENST00000344320.6
|
C15orf62
|
chromosome 15 open reading frame 62 |
| chr1_+_156084461 | 0.90 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr1_-_12679171 | 0.84 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr10_+_17270214 | 0.81 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr16_+_83986827 | 0.78 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr19_-_56056888 | 0.73 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr3_+_57094469 | 0.71 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr1_+_156096336 | 0.70 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr3_+_156807663 | 0.70 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr1_-_115880852 | 0.67 |
ENST00000369512.2
|
NGF
|
nerve growth factor (beta polypeptide) |
| chr14_+_38065052 | 0.66 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr4_-_185776854 | 0.64 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr2_-_96926313 | 0.63 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr9_-_34662651 | 0.62 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr3_-_196065248 | 0.61 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr9_+_131902346 | 0.61 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr11_-_62323702 | 0.59 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr4_+_159727272 | 0.58 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr17_+_21191341 | 0.58 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr15_+_91416092 | 0.58 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr6_+_151358048 | 0.57 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr1_-_6662919 | 0.56 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr12_-_57914275 | 0.55 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr9_-_91793675 | 0.54 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr1_-_6659876 | 0.53 |
ENST00000496707.1
|
KLHL21
|
kelch-like family member 21 |
| chr18_-_74207146 | 0.52 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr5_+_35852797 | 0.52 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr16_+_28996416 | 0.50 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr17_+_80317121 | 0.49 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr10_+_24755416 | 0.49 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_-_119066545 | 0.48 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr11_+_394196 | 0.48 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr15_-_67439270 | 0.48 |
ENST00000558463.1
|
RP11-342M21.2
|
Uncharacterized protein |
| chr7_+_102191679 | 0.47 |
ENST00000507918.1
ENST00000432940.1 |
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chr4_+_22999152 | 0.47 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr17_-_42994283 | 0.47 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr1_+_223889310 | 0.47 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr2_+_74741569 | 0.47 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr21_-_36421401 | 0.46 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr11_+_46316677 | 0.46 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr10_-_375422 | 0.45 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr11_-_8739383 | 0.44 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr5_+_149877440 | 0.44 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr17_+_74261413 | 0.44 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr17_-_74489215 | 0.44 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr16_-_1429674 | 0.44 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr19_+_7953417 | 0.44 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr9_+_140119618 | 0.43 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chr16_+_28875126 | 0.42 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr2_+_87755054 | 0.42 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_-_45272951 | 0.42 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr19_+_50431959 | 0.41 |
ENST00000595125.1
|
ATF5
|
activating transcription factor 5 |
| chr3_-_46608010 | 0.41 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr17_+_75315654 | 0.41 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr11_-_57417405 | 0.40 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr16_-_56223480 | 0.40 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr15_-_72521017 | 0.40 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr12_+_10365082 | 0.40 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_6341724 | 0.39 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr4_+_159727222 | 0.39 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr1_+_27189631 | 0.39 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr1_-_179112189 | 0.39 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chrX_-_48937531 | 0.39 |
ENST00000473974.1
ENST00000475880.1 ENST00000396681.4 ENST00000553851.1 ENST00000471338.1 ENST00000476728.1 ENST00000376368.2 ENST00000485908.1 ENST00000376372.3 ENST00000376358.3 |
WDR45
AF196779.12
|
WD repeat domain 45 WD repeat domain phosphoinositide-interacting protein 4 |
| chr7_+_55177416 | 0.39 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr1_+_156095951 | 0.38 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr16_-_1429627 | 0.38 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr7_+_150065278 | 0.38 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr17_-_27405875 | 0.38 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr18_+_21452804 | 0.38 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr11_+_35639735 | 0.38 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr11_-_2924720 | 0.37 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr19_+_50432400 | 0.37 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr1_+_39670423 | 0.37 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr1_-_153513170 | 0.36 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr7_+_73245193 | 0.36 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr3_+_174158732 | 0.36 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr17_+_74261277 | 0.36 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chrX_-_48937684 | 0.36 |
ENST00000465382.1
ENST00000423215.2 |
WDR45
|
WD repeat domain 45 |
| chr2_-_65593784 | 0.35 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr16_+_30077098 | 0.35 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_28875268 | 0.35 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr3_+_14474178 | 0.35 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr16_+_30418910 | 0.35 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr8_-_123139423 | 0.34 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr2_-_220083076 | 0.34 |
ENST00000295750.4
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr3_+_11267691 | 0.34 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr12_+_10365404 | 0.34 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_6341844 | 0.34 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr1_-_219615984 | 0.34 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr1_+_39670360 | 0.33 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr9_-_97356075 | 0.33 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr17_+_75315534 | 0.33 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr11_-_65430554 | 0.33 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr12_+_57914481 | 0.33 |
ENST00000548887.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr8_+_22844995 | 0.33 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr3_-_122512619 | 0.32 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr5_+_175288631 | 0.32 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_+_36621174 | 0.32 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr1_-_179112173 | 0.32 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr5_+_38148582 | 0.32 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr2_+_87808725 | 0.32 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr17_+_79650962 | 0.32 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr8_+_22844913 | 0.32 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr14_+_69726656 | 0.31 |
ENST00000337827.4
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr5_+_149877334 | 0.31 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr11_+_394145 | 0.31 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chrX_-_48901012 | 0.31 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr1_+_155107820 | 0.31 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr3_-_87040259 | 0.31 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr1_-_223853425 | 0.31 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr5_+_179247759 | 0.30 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr4_-_122085469 | 0.30 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr19_-_6057282 | 0.30 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_+_151791074 | 0.30 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr17_+_73780852 | 0.30 |
ENST00000589666.1
|
UNK
|
unkempt family zinc finger |
| chr19_+_38880695 | 0.30 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_+_6411636 | 0.30 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr1_+_174844645 | 0.29 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_+_77425972 | 0.29 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr20_+_48909240 | 0.29 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr16_-_4665023 | 0.29 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr1_+_12042015 | 0.29 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chrX_-_153285395 | 0.29 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chrX_-_153285251 | 0.29 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr11_-_75201791 | 0.29 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr4_-_987217 | 0.28 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr9_+_2015335 | 0.28 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_30685214 | 0.28 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr10_+_121410882 | 0.28 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr11_+_73000449 | 0.28 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_-_67141640 | 0.28 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr6_+_41604747 | 0.28 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr17_+_37030127 | 0.28 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr19_+_38794797 | 0.27 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr1_-_158656488 | 0.27 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr12_-_15114603 | 0.27 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr16_-_4164027 | 0.27 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr11_-_65430251 | 0.27 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr16_+_88519669 | 0.27 |
ENST00000319555.3
|
ZFPM1
|
zinc finger protein, FOG family member 1 |
| chr11_+_64126671 | 0.27 |
ENST00000530504.1
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr11_-_62457371 | 0.27 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr10_+_115312825 | 0.26 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr16_+_57662419 | 0.26 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr7_+_143078379 | 0.26 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr16_+_66968343 | 0.26 |
ENST00000417689.1
ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2
|
carboxylesterase 2 |
| chr9_+_131902283 | 0.26 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr8_-_38386175 | 0.26 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr19_+_38880252 | 0.26 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_59783941 | 0.25 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chrX_-_48937503 | 0.25 |
ENST00000322995.8
|
WDR45
|
WD repeat domain 45 |
| chr18_+_21452964 | 0.25 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr1_+_36621697 | 0.25 |
ENST00000373150.4
ENST00000373151.2 |
MAP7D1
|
MAP7 domain containing 1 |
| chr16_-_4664860 | 0.25 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr5_-_41794313 | 0.25 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr2_-_220117867 | 0.25 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr12_-_48214345 | 0.25 |
ENST00000433685.1
ENST00000447463.1 ENST00000427332.2 |
HDAC7
|
histone deacetylase 7 |
| chr6_+_25652501 | 0.24 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr18_-_25616519 | 0.24 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_-_8000872 | 0.24 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr12_-_91573132 | 0.24 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chrX_+_22056165 | 0.24 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr9_-_130341268 | 0.24 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr19_+_3366547 | 0.24 |
ENST00000341919.3
ENST00000590282.1 ENST00000443272.2 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr7_+_28448995 | 0.24 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr7_-_229557 | 0.24 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr16_+_30077055 | 0.24 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr19_+_36630454 | 0.24 |
ENST00000246533.3
|
CAPNS1
|
calpain, small subunit 1 |
| chr21_+_27543175 | 0.24 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr7_-_155604967 | 0.24 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr2_-_61389168 | 0.24 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr10_+_104155450 | 0.24 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr21_+_38792602 | 0.24 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr1_+_36621529 | 0.23 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr1_+_33722080 | 0.23 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr17_-_18908040 | 0.23 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr1_+_110693103 | 0.23 |
ENST00000331565.4
|
SLC6A17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr11_-_9781068 | 0.23 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr6_+_106546808 | 0.23 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr9_+_124329336 | 0.23 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr3_+_184016986 | 0.23 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr7_+_134528635 | 0.23 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr14_-_74226961 | 0.23 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr1_-_223853348 | 0.23 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr3_-_48632593 | 0.23 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr11_-_65667997 | 0.23 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr1_-_159894319 | 0.23 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr6_-_44233361 | 0.23 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr1_-_95007193 | 0.23 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr7_+_102290772 | 0.23 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr1_+_15943995 | 0.22 |
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr11_-_93271058 | 0.22 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr17_-_79881408 | 0.22 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr12_+_120972606 | 0.22 |
ENST00000413266.2
|
RNF10
|
ring finger protein 10 |
| chr10_+_123872483 | 0.22 |
ENST00000369001.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_108746282 | 0.22 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr10_-_22292675 | 0.22 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr12_-_111926342 | 0.22 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr17_+_75446819 | 0.22 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.2 | 0.6 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.2 | 0.6 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.2 | 0.7 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.6 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.3 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.3 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.3 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.8 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 1.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.2 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.6 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.8 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 1.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.5 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.5 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.2 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.4 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 1.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.0 | GO:0060364 | forebrain radial glial cell differentiation(GO:0021861) frontal suture morphogenesis(GO:0060364) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.5 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.4 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.1 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.7 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.2 | 2.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.6 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.7 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.8 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.3 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0005165 | neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0050659 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |