Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
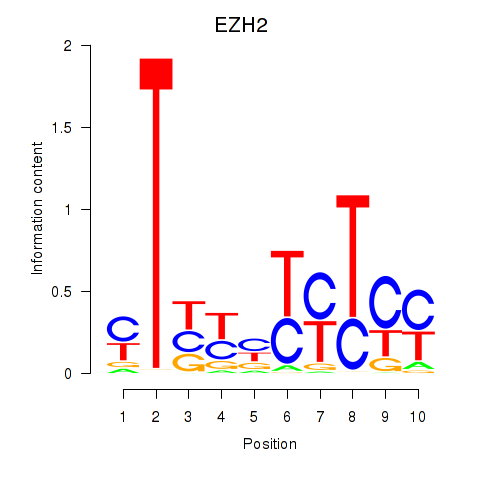
Results for EZH2
Z-value: 2.40
Transcription factors associated with EZH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EZH2
|
ENSG00000106462.6 | enhancer of zeste 2 polycomb repressive complex 2 subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EZH2 | hg19_v2_chr7_-_148581360_148581425 | 0.38 | 4.5e-01 | Click! |
Activity profile of EZH2 motif
Sorted Z-values of EZH2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_92919043 | 2.50 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_-_170151603 | 1.83 |
ENST00000366774.3
|
TCTE3
|
t-complex-associated-testis-expressed 3 |
| chr2_+_11864458 | 1.68 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr5_+_175298674 | 1.67 |
ENST00000514150.1
|
CPLX2
|
complexin 2 |
| chr5_+_175298573 | 1.59 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr1_+_95975672 | 1.56 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr19_-_4338783 | 1.51 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr11_+_6226782 | 1.51 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr17_-_46692287 | 1.45 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr5_+_158527630 | 1.41 |
ENST00000523301.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr6_+_144665237 | 1.31 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr2_+_45168875 | 1.30 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr5_+_175298487 | 1.26 |
ENST00000393745.3
|
CPLX2
|
complexin 2 |
| chr15_+_91416092 | 1.25 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr16_+_70207686 | 1.19 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr17_+_7387677 | 1.16 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr11_-_66496655 | 1.15 |
ENST00000527010.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr8_+_61592073 | 1.15 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr14_+_23352374 | 1.13 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr2_-_27435634 | 1.08 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr6_+_31540056 | 1.07 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr17_+_36584662 | 1.06 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr16_-_30569584 | 1.06 |
ENST00000252797.2
ENST00000568114.1 |
ZNF764
AC002310.13
|
zinc finger protein 764 Uncharacterized protein |
| chr3_+_193853927 | 1.06 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr13_+_51913819 | 1.05 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr19_-_56056888 | 1.04 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr3_+_169629354 | 1.02 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr14_+_59100774 | 1.01 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr14_+_23305760 | 1.00 |
ENST00000311852.6
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr14_+_23305783 | 0.99 |
ENST00000547279.1
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr11_-_8832182 | 0.95 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr6_-_121655850 | 0.93 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr7_+_69064566 | 0.92 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr5_-_158526756 | 0.91 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr7_-_155604967 | 0.90 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr16_+_30075783 | 0.89 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr17_+_75316336 | 0.89 |
ENST00000591934.1
|
SEPT9
|
septin 9 |
| chr17_-_7832753 | 0.89 |
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr1_+_39734131 | 0.88 |
ENST00000530262.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr22_-_31688431 | 0.88 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr20_-_62168714 | 0.87 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr1_+_202385953 | 0.87 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_-_89785777 | 0.87 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr17_-_48278983 | 0.87 |
ENST00000225964.5
|
COL1A1
|
collagen, type I, alpha 1 |
| chr20_-_42815733 | 0.87 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr7_-_41740181 | 0.86 |
ENST00000442711.1
|
INHBA
|
inhibin, beta A |
| chr11_-_66056596 | 0.86 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr17_+_7387919 | 0.84 |
ENST00000572844.1
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr17_+_55183261 | 0.83 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr11_-_66496430 | 0.83 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr11_-_44331679 | 0.83 |
ENST00000329255.3
|
ALX4
|
ALX homeobox 4 |
| chr19_+_51728316 | 0.82 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr1_+_26758790 | 0.81 |
ENST00000427245.2
ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr16_+_30662050 | 0.80 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr7_+_69064300 | 0.80 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr20_-_52612705 | 0.79 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr10_-_81203972 | 0.78 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr20_-_62168672 | 0.78 |
ENST00000217185.2
|
PTK6
|
protein tyrosine kinase 6 |
| chr5_+_158527485 | 0.76 |
ENST00000517335.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr17_-_36884451 | 0.76 |
ENST00000595377.1
|
AC006449.1
|
NS5ATP13TP1; Uncharacterized protein |
| chr16_+_31085714 | 0.76 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr4_+_2794750 | 0.75 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr12_-_70093235 | 0.74 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr17_+_7836398 | 0.74 |
ENST00000565740.1
|
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr3_-_9291063 | 0.74 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr15_+_63050785 | 0.73 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr15_-_88799948 | 0.73 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr8_-_7274385 | 0.73 |
ENST00000318157.2
|
DEFB4B
|
defensin, beta 4B |
| chrX_+_133941218 | 0.73 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr16_+_30075595 | 0.72 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_+_57105991 | 0.72 |
ENST00000263314.2
|
P2RX3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr8_+_58055238 | 0.71 |
ENST00000519314.1
ENST00000519241.1 |
RP11-513O17.2
|
RP11-513O17.2 |
| chr18_+_72167096 | 0.71 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr11_+_64004888 | 0.71 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr16_+_29127282 | 0.71 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr14_+_56584414 | 0.70 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_-_21289627 | 0.70 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr12_-_49418407 | 0.70 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr9_-_85882145 | 0.70 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr17_-_7387524 | 0.69 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr13_-_72441315 | 0.69 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr11_+_73676281 | 0.69 |
ENST00000543947.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr11_+_12302492 | 0.69 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr16_-_73093597 | 0.69 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr2_+_220436917 | 0.69 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr18_+_72166564 | 0.68 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr8_+_26435915 | 0.68 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr22_-_36236265 | 0.68 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr4_+_183065793 | 0.68 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr16_+_30709530 | 0.68 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr6_+_29624758 | 0.68 |
ENST00000376917.3
ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr20_-_656437 | 0.67 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chr1_+_36621174 | 0.67 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_-_40575535 | 0.67 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr6_-_10415218 | 0.66 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr1_+_156211753 | 0.66 |
ENST00000368272.4
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein |
| chr3_-_197300194 | 0.66 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr19_-_12992244 | 0.66 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr9_-_134585221 | 0.65 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_+_59783941 | 0.64 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr8_+_22857048 | 0.63 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr1_+_39670360 | 0.63 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr5_+_140749803 | 0.62 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr6_+_159071015 | 0.62 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr19_-_46088068 | 0.62 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr11_+_76092353 | 0.62 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr16_+_30662085 | 0.61 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr2_+_219472488 | 0.61 |
ENST00000450993.2
|
PLCD4
|
phospholipase C, delta 4 |
| chrX_+_133941250 | 0.61 |
ENST00000445123.1
|
FAM122C
|
family with sequence similarity 122C |
| chr17_-_47045949 | 0.61 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr11_-_126870655 | 0.61 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr12_+_58087901 | 0.61 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr6_+_30848829 | 0.60 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chrX_-_100129320 | 0.60 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr8_-_623547 | 0.60 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr12_+_66218212 | 0.60 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr10_-_77161533 | 0.60 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr12_+_10331605 | 0.60 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr6_-_37665751 | 0.60 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr16_+_31483451 | 0.59 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr11_+_19799327 | 0.59 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr16_-_2260834 | 0.59 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr11_-_62414070 | 0.59 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chrY_+_15418467 | 0.59 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr3_+_42190714 | 0.58 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr3_-_121379739 | 0.58 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr3_+_111260980 | 0.58 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr17_-_39538550 | 0.58 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr7_-_130597935 | 0.58 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr1_-_151319318 | 0.58 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr3_+_50654821 | 0.58 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr15_+_84908573 | 0.58 |
ENST00000424966.1
ENST00000422563.2 |
GOLGA6L4
|
golgin A6 family-like 4 |
| chr1_-_72748417 | 0.58 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr2_+_63277927 | 0.58 |
ENST00000282549.2
|
OTX1
|
orthodenticle homeobox 1 |
| chr4_+_160188889 | 0.57 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr17_+_4736812 | 0.57 |
ENST00000453408.3
|
MINK1
|
misshapen-like kinase 1 |
| chr6_-_131277510 | 0.57 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr17_+_45286706 | 0.57 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr1_+_206557366 | 0.56 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr15_+_63335899 | 0.56 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr6_+_148593425 | 0.56 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr8_-_21646311 | 0.55 |
ENST00000524240.1
ENST00000400782.4 |
GFRA2
|
GDNF family receptor alpha 2 |
| chr17_-_74489215 | 0.55 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chrX_+_48681768 | 0.55 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr16_-_19725899 | 0.55 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr6_+_158733692 | 0.54 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr3_+_10312604 | 0.54 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr2_-_69870835 | 0.54 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr12_-_88423164 | 0.54 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr16_+_28996572 | 0.54 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr2_-_220436248 | 0.53 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr17_+_79953310 | 0.53 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr17_-_2117600 | 0.53 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr11_-_64014379 | 0.53 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_61596753 | 0.53 |
ENST00000448607.1
ENST00000421879.1 |
FADS1
|
fatty acid desaturase 1 |
| chr4_-_46911223 | 0.53 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr12_-_118406777 | 0.52 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr12_-_118406028 | 0.52 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr1_-_154941350 | 0.52 |
ENST00000444179.1
ENST00000414115.1 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr12_-_49449107 | 0.52 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr9_+_131902283 | 0.52 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr10_-_75682535 | 0.52 |
ENST00000409178.1
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr15_-_89438742 | 0.52 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr1_-_153588765 | 0.52 |
ENST00000368701.1
ENST00000344616.2 |
S100A14
|
S100 calcium binding protein A14 |
| chr2_-_98950434 | 0.52 |
ENST00000409490.1
|
AC092675.3
|
Uncharacterized protein |
| chr16_-_67281413 | 0.51 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr1_-_46598284 | 0.51 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_+_156589198 | 0.51 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr9_-_128246769 | 0.51 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr4_-_80994619 | 0.51 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr6_+_42952237 | 0.51 |
ENST00000485511.1
ENST00000394110.3 ENST00000472118.1 ENST00000461010.1 |
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta |
| chr12_-_115121962 | 0.51 |
ENST00000349155.2
|
TBX3
|
T-box 3 |
| chr4_+_110749143 | 0.51 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr9_+_104296163 | 0.51 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chrX_-_19688475 | 0.51 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr19_-_54604083 | 0.51 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr1_+_156052354 | 0.51 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr11_-_119066545 | 0.51 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr12_+_2912363 | 0.50 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr11_+_118401899 | 0.50 |
ENST00000528373.1
ENST00000544878.1 ENST00000354284.4 ENST00000533137.1 ENST00000532762.1 ENST00000526973.1 ENST00000354064.7 ENST00000533102.1 ENST00000313236.5 ENST00000527267.1 ENST00000524725.1 ENST00000533689.1 |
TMEM25
|
transmembrane protein 25 |
| chr22_+_42017987 | 0.50 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr17_+_27895045 | 0.50 |
ENST00000580183.2
ENST00000578749.1 ENST00000582829.2 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr12_-_116714564 | 0.49 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr1_+_18807424 | 0.49 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr1_+_36621529 | 0.49 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr16_-_31085514 | 0.49 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr10_-_16563870 | 0.48 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr6_+_30848771 | 0.48 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_+_100271355 | 0.48 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr2_-_190044480 | 0.48 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr5_+_102200948 | 0.48 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_+_100187196 | 0.48 |
ENST00000468962.1
ENST00000427939.2 |
FBXO24
|
F-box protein 24 |
| chr18_-_56296182 | 0.48 |
ENST00000361673.3
|
ALPK2
|
alpha-kinase 2 |
| chr6_-_127837739 | 0.48 |
ENST00000368268.2
|
SOGA3
|
SOGA family member 3 |
| chr15_+_22368478 | 0.47 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr1_+_156589051 | 0.47 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr9_-_107690420 | 0.47 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr14_+_24590560 | 0.47 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr7_-_100881041 | 0.47 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr12_+_54426637 | 0.47 |
ENST00000312492.2
|
HOXC5
|
homeobox C5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EZH2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.4 | 0.9 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 1.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 1.7 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 1.3 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.4 | 1.6 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.4 | 1.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.3 | 1.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 0.9 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.2 | 1.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.9 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 0.7 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.2 | 2.0 | GO:1990834 | response to odorant(GO:1990834) |
| 0.2 | 0.6 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 0.8 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 0.6 | GO:0061114 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.6 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 1.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.2 | 0.8 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.2 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.2 | 0.6 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 0.9 | GO:0021622 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.2 | 0.7 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 1.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 0.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.2 | 1.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.2 | 1.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.2 | 4.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.5 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.2 | 0.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 1.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.7 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 1.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.6 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.5 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 2.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.3 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.7 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.1 | 0.4 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.4 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.7 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.6 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 | 0.4 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.3 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.3 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 1.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.0 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.3 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 1.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 1.9 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.7 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.9 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.4 | GO:0003284 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.4 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.1 | GO:0070647 | protein modification by small protein conjugation or removal(GO:0070647) |
| 0.1 | 0.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.6 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.3 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 1.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.5 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.6 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.1 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.2 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.4 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 1.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.5 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.7 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.4 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.5 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.7 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.1 | GO:0051495 | positive regulation of cytoskeleton organization(GO:0051495) |
| 0.1 | 0.4 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.5 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.4 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) protein secretion by platelet(GO:0070560) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.4 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.2 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.7 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.2 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.6 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 1.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 1.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 1.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.5 | GO:0006189 | brainstem development(GO:0003360) 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.0 | 0.5 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 1.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 1.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.4 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.3 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.5 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 1.1 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 1.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 1.6 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.4 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.3 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.3 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0060525 | prostate epithelial cord elongation(GO:0060523) prostate glandular acinus development(GO:0060525) prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.0 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0070570 | regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.0 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.4 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 1.1 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:1900747 | xanthine metabolic process(GO:0046110) negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.9 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.3 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.4 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.6 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.4 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 1.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 1.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 4.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.2 | 0.2 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.2 | 1.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.8 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.2 | 2.0 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.6 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 1.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.3 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.3 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.8 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 2.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.3 | GO:0016939 | plus-end kinesin complex(GO:0005873) kinesin II complex(GO:0016939) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.2 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.3 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 1.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.4 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.7 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.2 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 2.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.9 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.6 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.4 | 1.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 2.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 1.4 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 0.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 0.8 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 2.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.6 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 0.5 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.2 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.9 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.4 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 1.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.5 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.3 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 1.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.4 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.4 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.7 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.5 | GO:0044212 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 3.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.0 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.0 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.0 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |