Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
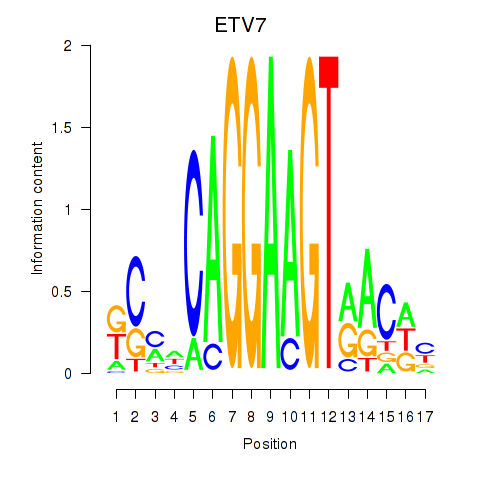
Results for ETV7
Z-value: 0.59
Transcription factors associated with ETV7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV7
|
ENSG00000010030.9 | ETS variant transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV7 | hg19_v2_chr6_-_36355486_36355497 | 0.79 | 6.2e-02 | Click! |
Activity profile of ETV7 motif
Sorted Z-values of ETV7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_19281203 | 0.33 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr1_-_1590418 | 0.32 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr10_-_18948208 | 0.29 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr6_-_26235206 | 0.29 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr16_-_75467274 | 0.21 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr7_+_1609765 | 0.19 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr12_+_6561190 | 0.19 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr22_-_36877371 | 0.18 |
ENST00000403313.1
|
TXN2
|
thioredoxin 2 |
| chr9_+_134001455 | 0.18 |
ENST00000531584.1
|
NUP214
|
nucleoporin 214kDa |
| chr5_-_72861175 | 0.18 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr3_-_49933249 | 0.18 |
ENST00000434765.1
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr12_+_72058130 | 0.18 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr22_+_38035623 | 0.17 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr19_-_4791219 | 0.17 |
ENST00000598782.1
|
AC005523.3
|
AC005523.3 |
| chr2_+_242716231 | 0.17 |
ENST00000192314.6
|
GAL3ST2
|
galactose-3-O-sulfotransferase 2 |
| chr1_+_37947257 | 0.17 |
ENST00000471012.1
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr14_-_90097910 | 0.16 |
ENST00000550332.2
|
RP11-944C7.1
|
Protein LOC100506792 |
| chr17_+_7482785 | 0.15 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr3_+_97483366 | 0.15 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr10_+_88718397 | 0.15 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr3_-_170459154 | 0.15 |
ENST00000473110.1
|
RP11-373E16.4
|
RP11-373E16.4 |
| chr17_+_19281034 | 0.15 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr9_-_139891165 | 0.15 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr1_-_152552980 | 0.15 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr17_-_42188598 | 0.14 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr2_-_200715573 | 0.14 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr21_-_46340884 | 0.14 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr16_-_4401284 | 0.14 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr5_-_79866054 | 0.13 |
ENST00000508916.1
|
ANKRD34B
|
ankyrin repeat domain 34B |
| chr15_+_23810853 | 0.13 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr9_+_134000948 | 0.13 |
ENST00000359428.5
ENST00000411637.2 ENST00000451030.1 |
NUP214
|
nucleoporin 214kDa |
| chr1_-_1655713 | 0.13 |
ENST00000401096.2
ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A
|
cyclin-dependent kinase 11A |
| chr22_+_27068766 | 0.13 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr22_-_37640456 | 0.12 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr7_+_1609694 | 0.12 |
ENST00000437621.2
ENST00000457484.2 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr17_+_78193443 | 0.12 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr2_+_9696028 | 0.11 |
ENST00000607241.1
|
RP11-214N9.1
|
RP11-214N9.1 |
| chr1_+_203734296 | 0.11 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr10_+_47658234 | 0.11 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr1_+_156698743 | 0.11 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr7_-_5821225 | 0.11 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr6_-_33267101 | 0.11 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr5_+_180650271 | 0.11 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr12_-_69080590 | 0.11 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr15_-_72462384 | 0.11 |
ENST00000568594.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr17_+_9479944 | 0.11 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr3_-_121379739 | 0.11 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr20_-_60294804 | 0.10 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr9_-_100000957 | 0.10 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr1_-_249120832 | 0.10 |
ENST00000366472.5
|
SH3BP5L
|
SH3-binding domain protein 5-like |
| chr17_-_7082861 | 0.10 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr4_-_183838596 | 0.10 |
ENST00000508994.1
ENST00000512766.1 |
DCTD
|
dCMP deaminase |
| chr19_+_17862274 | 0.10 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chr20_+_35918035 | 0.10 |
ENST00000373606.3
ENST00000397156.3 ENST00000397150.1 ENST00000397152.3 |
MANBAL
|
mannosidase, beta A, lysosomal-like |
| chr11_-_57102947 | 0.10 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr3_-_47324060 | 0.10 |
ENST00000452770.2
|
KIF9
|
kinesin family member 9 |
| chr2_+_238969530 | 0.10 |
ENST00000254663.6
ENST00000555827.1 ENST00000373332.3 ENST00000413463.1 ENST00000409736.2 ENST00000422984.2 ENST00000412508.1 ENST00000429612.2 |
SCLY
|
selenocysteine lyase |
| chr11_+_64008525 | 0.10 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr17_+_80193644 | 0.09 |
ENST00000582946.1
|
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_-_150563 | 0.09 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr13_-_46626820 | 0.09 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr2_-_97534312 | 0.09 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr20_+_60758075 | 0.09 |
ENST00000536470.1
ENST00000436421.2 ENST00000370823.3 ENST00000448254.1 |
MTG2
|
mitochondrial ribosome-associated GTPase 2 |
| chr22_+_37678424 | 0.09 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chr11_+_64008443 | 0.09 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_-_226111929 | 0.09 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr9_-_134151915 | 0.09 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr15_-_80263506 | 0.09 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr1_+_150122034 | 0.09 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr12_+_123464607 | 0.09 |
ENST00000543566.1
ENST00000315580.5 ENST00000542099.1 ENST00000392435.2 ENST00000413381.2 ENST00000426960.2 ENST00000453766.2 |
ARL6IP4
|
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr6_-_33385870 | 0.09 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr19_+_53073526 | 0.09 |
ENST00000596514.1
ENST00000391785.3 ENST00000301093.2 |
ZNF701
|
zinc finger protein 701 |
| chr1_+_156024552 | 0.09 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr5_+_61708488 | 0.09 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr17_-_74722536 | 0.08 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chr17_+_21030260 | 0.08 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr4_+_87928353 | 0.08 |
ENST00000511722.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr16_-_71323617 | 0.08 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr4_+_24661479 | 0.08 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr22_+_21369316 | 0.08 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr16_-_66968055 | 0.08 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr22_+_50312379 | 0.08 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr19_-_2090131 | 0.08 |
ENST00000591326.1
|
MOB3A
|
MOB kinase activator 3A |
| chr22_-_36925186 | 0.08 |
ENST00000541106.1
ENST00000455547.1 ENST00000432675.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr12_+_9066472 | 0.08 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr17_-_79849438 | 0.08 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr6_+_31515337 | 0.08 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr8_+_145133493 | 0.08 |
ENST00000316052.5
ENST00000525936.1 |
EXOSC4
|
exosome component 4 |
| chr16_+_1832902 | 0.08 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr22_-_36924944 | 0.08 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr6_-_47445214 | 0.08 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr22_-_36877629 | 0.08 |
ENST00000416967.1
|
TXN2
|
thioredoxin 2 |
| chr17_-_7218631 | 0.08 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr20_-_48532019 | 0.08 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr12_-_53297432 | 0.08 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr18_+_43246028 | 0.08 |
ENST00000589658.1
|
SLC14A2
|
solute carrier family 14 (urea transporter), member 2 |
| chr15_-_65117807 | 0.08 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr10_+_75668916 | 0.08 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr12_+_81471816 | 0.08 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr12_+_123459127 | 0.08 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr5_-_180237082 | 0.08 |
ENST00000506889.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr6_-_31648150 | 0.08 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr22_+_38035459 | 0.08 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr19_-_36236292 | 0.08 |
ENST00000378975.3
ENST00000412391.2 ENST00000292879.5 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr16_+_30078811 | 0.07 |
ENST00000564688.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_+_19988736 | 0.07 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr9_-_131085021 | 0.07 |
ENST00000372890.4
|
TRUB2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr11_-_6502580 | 0.07 |
ENST00000423813.2
ENST00000396777.3 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr16_-_87799505 | 0.07 |
ENST00000353170.5
ENST00000561825.1 ENST00000270583.5 ENST00000562261.1 ENST00000347925.5 |
KLHDC4
|
kelch domain containing 4 |
| chr19_-_11039261 | 0.07 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr3_-_145940214 | 0.07 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr14_-_67878917 | 0.07 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr7_-_130597935 | 0.07 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr2_-_231084820 | 0.07 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr17_+_75315654 | 0.07 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr19_+_55996543 | 0.07 |
ENST00000591590.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr16_-_734318 | 0.07 |
ENST00000609261.1
ENST00000562111.1 ENST00000562824.1 ENST00000412368.2 ENST00000293882.4 ENST00000454700.1 |
JMJD8
|
jumonji domain containing 8 |
| chr1_-_27480973 | 0.07 |
ENST00000545949.1
ENST00000374086.3 |
SLC9A1
|
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr19_-_10230540 | 0.07 |
ENST00000589454.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr22_+_27068704 | 0.07 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr3_-_49893907 | 0.07 |
ENST00000482582.1
|
TRAIP
|
TRAF interacting protein |
| chr14_-_57197224 | 0.07 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr5_+_140213815 | 0.07 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr12_+_38710555 | 0.07 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr7_-_100888337 | 0.07 |
ENST00000223136.4
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr2_+_217363559 | 0.07 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr17_-_8198636 | 0.07 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr12_-_52604607 | 0.07 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr6_+_30951487 | 0.07 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr17_-_80231557 | 0.07 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chrX_-_48901012 | 0.07 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr16_+_23652773 | 0.07 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr22_+_30805086 | 0.07 |
ENST00000439838.1
ENST00000439023.3 |
RP4-539M6.19
|
Uncharacterized protein |
| chr3_-_53080047 | 0.06 |
ENST00000482396.1
ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr4_+_79567362 | 0.06 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr20_-_33872548 | 0.06 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr17_-_7155802 | 0.06 |
ENST00000572043.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr22_-_37640277 | 0.06 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr19_-_53238260 | 0.06 |
ENST00000453741.2
ENST00000602162.1 ENST00000601643.1 ENST00000596702.1 ENST00000600943.1 ENST00000543227.1 ENST00000540744.1 |
ZNF611
|
zinc finger protein 611 |
| chr9_-_77703115 | 0.06 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr7_-_102105289 | 0.06 |
ENST00000292566.3
|
ALKBH4
|
alkB, alkylation repair homolog 4 (E. coli) |
| chr2_+_87808725 | 0.06 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr22_+_18560743 | 0.06 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr14_+_24563262 | 0.06 |
ENST00000559250.1
ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr7_-_5553369 | 0.06 |
ENST00000453700.3
ENST00000382368.3 |
FBXL18
|
F-box and leucine-rich repeat protein 18 |
| chr2_+_175260514 | 0.06 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr17_+_7835419 | 0.06 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr22_-_36925124 | 0.06 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr1_+_32666188 | 0.06 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr10_-_43187153 | 0.06 |
ENST00000411439.1
|
AL022344.4
|
AL022344.4 |
| chr21_-_46340807 | 0.06 |
ENST00000397846.3
ENST00000524251.1 ENST00000522688.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_+_11039391 | 0.06 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr19_-_3500635 | 0.06 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr6_-_33385902 | 0.06 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr8_+_67104323 | 0.06 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr8_-_82754193 | 0.06 |
ENST00000519817.1
ENST00000521773.1 ENST00000523757.1 |
SNX16
|
sorting nexin 16 |
| chr5_-_110074603 | 0.06 |
ENST00000515278.2
|
TMEM232
|
transmembrane protein 232 |
| chr1_+_43855545 | 0.06 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr17_-_74722672 | 0.06 |
ENST00000397625.4
ENST00000445478.2 |
JMJD6
|
jumonji domain containing 6 |
| chr2_-_220041860 | 0.06 |
ENST00000453038.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr1_-_36020531 | 0.06 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr12_+_94071129 | 0.06 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_-_49893958 | 0.06 |
ENST00000482243.1
ENST00000331456.2 ENST00000469027.1 |
TRAIP
|
TRAF interacting protein |
| chr13_-_45048386 | 0.06 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr4_-_11431188 | 0.06 |
ENST00000510712.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr2_-_170681324 | 0.06 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chr17_+_40688190 | 0.06 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr9_-_130667592 | 0.06 |
ENST00000447681.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr17_-_76836963 | 0.06 |
ENST00000312010.6
|
USP36
|
ubiquitin specific peptidase 36 |
| chr3_-_25915189 | 0.06 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr16_+_77225071 | 0.06 |
ENST00000439557.2
ENST00000545553.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr16_-_66968265 | 0.06 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr14_+_24899141 | 0.06 |
ENST00000556842.1
ENST00000553935.1 |
KHNYN
|
KH and NYN domain containing |
| chr19_-_8675559 | 0.06 |
ENST00000597188.1
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr6_+_35704804 | 0.06 |
ENST00000373869.3
|
ARMC12
|
armadillo repeat containing 12 |
| chr19_+_36236491 | 0.06 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr22_-_29949680 | 0.06 |
ENST00000397873.2
ENST00000490103.1 |
THOC5
|
THO complex 5 |
| chr10_-_128110441 | 0.06 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr14_-_94595993 | 0.06 |
ENST00000238609.3
|
IFI27L2
|
interferon, alpha-inducible protein 27-like 2 |
| chr1_-_51791596 | 0.06 |
ENST00000532836.1
ENST00000422925.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr11_-_6502534 | 0.06 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr10_-_81742364 | 0.06 |
ENST00000444384.3
|
SFTPD
|
surfactant protein D |
| chr21_-_46340770 | 0.06 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_+_27648709 | 0.06 |
ENST00000608611.1
ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222
|
transmembrane protein 222 |
| chr3_-_47950745 | 0.06 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr8_-_7320974 | 0.06 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr15_-_72462705 | 0.06 |
ENST00000564129.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr14_+_55590646 | 0.06 |
ENST00000553493.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chrX_+_105066524 | 0.05 |
ENST00000243300.9
ENST00000428173.2 |
NRK
|
Nik related kinase |
| chr5_-_140070897 | 0.05 |
ENST00000448240.1
ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS
|
histidyl-tRNA synthetase |
| chr6_-_32095968 | 0.05 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr17_+_4981535 | 0.05 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr17_-_27916589 | 0.05 |
ENST00000579937.1
ENST00000335356.7 |
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr14_+_24563510 | 0.05 |
ENST00000545054.2
ENST00000561286.1 ENST00000558096.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr17_+_6918354 | 0.05 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr11_-_8739383 | 0.05 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr1_+_156698234 | 0.05 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_+_79849872 | 0.05 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr2_-_231084617 | 0.05 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr9_+_139873264 | 0.05 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.2 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.1 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.2 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.0 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.0 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.0 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.0 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |