Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
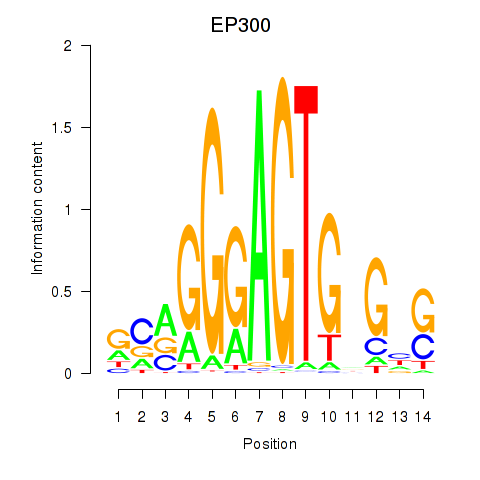
Results for EP300
Z-value: 1.23
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.9 | E1A binding protein p300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg19_v2_chr22_+_41487711_41487798 | 0.19 | 7.1e-01 | Click! |
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_20866424 | 1.11 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr13_-_44361025 | 0.60 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr1_-_32827682 | 0.57 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chr20_-_19738569 | 0.53 |
ENST00000598007.1
|
AL121761.2
|
Uncharacterized protein |
| chr11_+_46722368 | 0.52 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr17_+_43238438 | 0.51 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr3_-_126076264 | 0.47 |
ENST00000296233.3
|
KLF15
|
Kruppel-like factor 15 |
| chr8_-_37594944 | 0.47 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr15_+_23810853 | 0.43 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr16_-_31085033 | 0.42 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr10_+_81892477 | 0.40 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr17_+_79031415 | 0.40 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr1_+_32712815 | 0.39 |
ENST00000373582.3
|
FAM167B
|
family with sequence similarity 167, member B |
| chr15_-_93353028 | 0.36 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr3_-_118753792 | 0.35 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr15_-_74726283 | 0.34 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr6_-_30684898 | 0.34 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr1_+_116519223 | 0.34 |
ENST00000369502.1
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr11_+_2421718 | 0.33 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr9_-_91793675 | 0.33 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr12_+_104235229 | 0.33 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr19_-_44809121 | 0.31 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr1_-_1590418 | 0.31 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr11_+_46403194 | 0.31 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_+_2397402 | 0.31 |
ENST00000475945.2
|
CD81
|
CD81 molecule |
| chr2_+_220436917 | 0.30 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr5_-_443239 | 0.30 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chr8_+_144816303 | 0.29 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr12_-_105629852 | 0.29 |
ENST00000551662.1
ENST00000553097.1 |
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr22_-_29784519 | 0.28 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr11_-_46722117 | 0.28 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr12_+_70760056 | 0.28 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr19_-_1174226 | 0.28 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr15_+_42120283 | 0.28 |
ENST00000542534.2
ENST00000397299.4 ENST00000408047.1 ENST00000431823.1 ENST00000382448.4 ENST00000342159.4 |
PLA2G4B
JMJD7
JMJD7-PLA2G4B
|
phospholipase A2, group IVB (cytosolic) jumonji domain containing 7 JMJD7-PLA2G4B readthrough |
| chr16_+_69373471 | 0.28 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr1_+_206643787 | 0.27 |
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr13_-_40177261 | 0.27 |
ENST00000379589.3
|
LHFP
|
lipoma HMGIC fusion partner |
| chr18_+_5238055 | 0.27 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr22_-_30968813 | 0.27 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_+_11039391 | 0.27 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr5_+_443280 | 0.26 |
ENST00000508022.1
|
EXOC3
|
exocyst complex component 3 |
| chr8_+_27169138 | 0.26 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_-_209957882 | 0.25 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr1_-_161008697 | 0.25 |
ENST00000318289.10
ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr14_-_69445793 | 0.25 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr11_+_66234216 | 0.25 |
ENST00000349459.6
ENST00000320740.7 ENST00000524466.1 ENST00000526296.1 |
PELI3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr16_+_68279256 | 0.25 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr1_+_206643806 | 0.25 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr11_+_47430133 | 0.25 |
ENST00000531974.1
ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr19_+_10541462 | 0.25 |
ENST00000293683.5
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr18_-_5238525 | 0.25 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr14_-_69446034 | 0.24 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr17_-_74099795 | 0.24 |
ENST00000406660.3
ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7
|
exocyst complex component 7 |
| chr19_+_29704142 | 0.24 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr16_+_68279207 | 0.24 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr16_+_1832902 | 0.23 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr3_-_50649192 | 0.23 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr2_+_25015968 | 0.23 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr19_-_56109119 | 0.23 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr9_-_33473882 | 0.23 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr11_+_62623544 | 0.23 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr22_-_20104700 | 0.23 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr16_-_69373396 | 0.23 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr14_-_69445968 | 0.23 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr20_+_34020827 | 0.22 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr12_-_49365501 | 0.22 |
ENST00000403957.1
ENST00000301061.4 |
WNT10B
|
wingless-type MMTV integration site family, member 10B |
| chr14_+_56584414 | 0.22 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr10_+_30723533 | 0.22 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr16_-_31085514 | 0.22 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr12_-_57505121 | 0.22 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr21_-_46707793 | 0.22 |
ENST00000331343.7
ENST00000349485.5 |
POFUT2
|
protein O-fucosyltransferase 2 |
| chr20_-_30311703 | 0.22 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr11_+_62623512 | 0.22 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr17_-_79269067 | 0.21 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chr17_-_74099772 | 0.21 |
ENST00000411744.2
ENST00000332065.5 |
EXOC7
|
exocyst complex component 7 |
| chr11_+_62623621 | 0.21 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_-_6110474 | 0.21 |
ENST00000587181.1
ENST00000587321.1 ENST00000586806.1 ENST00000589742.1 ENST00000592546.1 ENST00000303657.5 |
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr1_-_95007193 | 0.21 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr17_+_4843679 | 0.21 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr20_+_44035200 | 0.21 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr9_-_77567743 | 0.21 |
ENST00000376854.5
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr17_-_76836963 | 0.20 |
ENST00000312010.6
|
USP36
|
ubiquitin specific peptidase 36 |
| chr19_-_49955050 | 0.20 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr19_+_51226573 | 0.20 |
ENST00000250340.4
|
CLEC11A
|
C-type lectin domain family 11, member A |
| chr16_+_66968343 | 0.20 |
ENST00000417689.1
ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2
|
carboxylesterase 2 |
| chr16_+_30205225 | 0.20 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr14_+_23340822 | 0.20 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr19_+_49617581 | 0.20 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr17_+_37844331 | 0.20 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr15_-_82338460 | 0.20 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr11_+_65819802 | 0.20 |
ENST00000528302.1
ENST00000322535.6 ENST00000524627.1 ENST00000533595.1 ENST00000530322.1 |
SF3B2
|
splicing factor 3b, subunit 2, 145kDa |
| chr5_+_443268 | 0.20 |
ENST00000512944.1
|
EXOC3
|
exocyst complex component 3 |
| chr16_+_4475806 | 0.20 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr19_-_49955096 | 0.20 |
ENST00000595550.1
|
PIH1D1
|
PIH1 domain containing 1 |
| chr17_-_56065540 | 0.20 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr19_-_18649181 | 0.20 |
ENST00000596015.1
|
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr11_-_107582775 | 0.19 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr16_+_29465822 | 0.19 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr6_-_41703296 | 0.19 |
ENST00000373033.1
|
TFEB
|
transcription factor EB |
| chr2_+_25016282 | 0.19 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr9_-_127703333 | 0.19 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr17_-_37844267 | 0.19 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr12_+_116997186 | 0.19 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr22_+_21321447 | 0.19 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr16_-_87799505 | 0.19 |
ENST00000353170.5
ENST00000561825.1 ENST00000270583.5 ENST00000562261.1 ENST00000347925.5 |
KLHDC4
|
kelch domain containing 4 |
| chr19_-_17186229 | 0.19 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr7_-_150974494 | 0.19 |
ENST00000392811.2
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr1_+_153700518 | 0.19 |
ENST00000318967.2
ENST00000456435.1 ENST00000435409.2 |
INTS3
|
integrator complex subunit 3 |
| chr12_-_6484376 | 0.18 |
ENST00000360168.3
ENST00000358945.3 |
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr6_+_43738444 | 0.18 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr19_+_17186577 | 0.18 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr8_-_134309335 | 0.18 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr19_-_45927097 | 0.18 |
ENST00000340192.7
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr12_+_52463751 | 0.18 |
ENST00000336854.4
ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44
|
chromosome 12 open reading frame 44 |
| chr19_+_10362882 | 0.18 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_-_2126174 | 0.18 |
ENST00000400919.3
ENST00000420515.1 ENST00000378543.2 ENST00000400918.3 |
C1orf86
|
chromosome 1 open reading frame 86 |
| chr16_-_30022735 | 0.18 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr1_-_33366931 | 0.18 |
ENST00000373463.3
ENST00000329151.5 |
TMEM54
|
transmembrane protein 54 |
| chr7_-_5569588 | 0.18 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr1_-_1655713 | 0.18 |
ENST00000401096.2
ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A
|
cyclin-dependent kinase 11A |
| chr1_+_32827759 | 0.18 |
ENST00000373534.3
|
TSSK3
|
testis-specific serine kinase 3 |
| chr9_+_130469257 | 0.18 |
ENST00000373295.2
|
C9orf117
|
chromosome 9 open reading frame 117 |
| chr9_+_133454943 | 0.18 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr16_+_77224732 | 0.17 |
ENST00000569610.1
ENST00000248248.3 ENST00000567291.1 ENST00000320859.6 ENST00000563612.1 ENST00000563279.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr11_+_46403303 | 0.17 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr22_-_19109901 | 0.17 |
ENST00000545799.1
ENST00000537045.1 ENST00000263196.7 |
DGCR2
|
DiGeorge syndrome critical region gene 2 |
| chr5_+_42423872 | 0.17 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr19_-_50083803 | 0.17 |
ENST00000391853.3
ENST00000339093.3 |
NOSIP
|
nitric oxide synthase interacting protein |
| chr1_-_40237020 | 0.17 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr19_+_37825526 | 0.17 |
ENST00000592768.1
ENST00000591417.1 ENST00000585623.1 ENST00000592168.1 ENST00000591391.1 ENST00000392153.3 ENST00000589392.1 ENST00000324411.4 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr16_+_69373323 | 0.17 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr15_+_74287009 | 0.17 |
ENST00000395135.3
|
PML
|
promyelocytic leukemia |
| chr19_+_10362577 | 0.17 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr7_-_139756791 | 0.17 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr4_+_8201091 | 0.16 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr1_+_157963391 | 0.16 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr18_+_5238549 | 0.16 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr6_+_43044003 | 0.16 |
ENST00000230419.4
ENST00000476760.1 ENST00000471863.1 ENST00000349241.2 ENST00000352931.2 ENST00000345201.2 |
PTK7
|
protein tyrosine kinase 7 |
| chr16_+_2732476 | 0.16 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr1_-_40367530 | 0.16 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_+_43855545 | 0.16 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr14_-_24711764 | 0.16 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr20_-_22565101 | 0.16 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr6_+_126240442 | 0.16 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_-_50083822 | 0.16 |
ENST00000596358.1
|
NOSIP
|
nitric oxide synthase interacting protein |
| chr1_-_16763685 | 0.16 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chrX_-_54522558 | 0.16 |
ENST00000375135.3
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr4_-_74853897 | 0.16 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr1_+_109656579 | 0.16 |
ENST00000526264.1
ENST00000369939.3 |
KIAA1324
|
KIAA1324 |
| chr3_+_19988736 | 0.16 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr17_-_7137582 | 0.15 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr10_-_104192405 | 0.15 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr20_+_47835884 | 0.15 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr17_-_71308119 | 0.15 |
ENST00000439510.2
ENST00000581014.1 ENST00000579611.1 |
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr16_+_718086 | 0.15 |
ENST00000315082.4
ENST00000563134.1 |
RHOT2
|
ras homolog family member T2 |
| chr2_+_201171372 | 0.15 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_19265982 | 0.15 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr19_-_4670345 | 0.15 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr10_+_102729249 | 0.15 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr1_-_2126192 | 0.15 |
ENST00000378546.4
|
C1orf86
|
chromosome 1 open reading frame 86 |
| chr19_+_11546093 | 0.15 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr17_+_40118805 | 0.15 |
ENST00000591072.1
ENST00000587679.1 ENST00000393888.1 ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr9_-_138799070 | 0.15 |
ENST00000389532.4
ENST00000409386.3 |
CAMSAP1
|
calmodulin regulated spectrin-associated protein 1 |
| chr6_-_34360413 | 0.15 |
ENST00000607016.1
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr20_-_33460621 | 0.15 |
ENST00000427420.1
ENST00000336431.5 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr19_-_16653226 | 0.15 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr17_+_73089382 | 0.15 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_-_94079648 | 0.15 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr22_+_37956479 | 0.15 |
ENST00000430687.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr15_+_74287035 | 0.15 |
ENST00000395132.2
ENST00000268059.6 ENST00000354026.6 ENST00000268058.3 ENST00000565898.1 ENST00000569477.1 ENST00000569965.1 ENST00000567543.1 ENST00000436891.3 ENST00000435786.2 ENST00000564428.1 ENST00000359928.4 |
PML
|
promyelocytic leukemia |
| chr3_+_57741957 | 0.15 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr20_-_33735070 | 0.15 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr16_+_77225071 | 0.15 |
ENST00000439557.2
ENST00000545553.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr17_+_47075023 | 0.15 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr17_+_4736627 | 0.15 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr8_+_30300119 | 0.14 |
ENST00000520191.1
|
RBPMS
|
RNA binding protein with multiple splicing |
| chr1_+_39734131 | 0.14 |
ENST00000530262.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr12_+_121416489 | 0.14 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr12_+_53693466 | 0.14 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr2_+_87808725 | 0.14 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_-_19229014 | 0.14 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr5_+_154136702 | 0.14 |
ENST00000524248.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr22_-_29075853 | 0.14 |
ENST00000397906.2
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr3_-_197463590 | 0.14 |
ENST00000296343.5
ENST00000389665.5 ENST00000449205.1 |
KIAA0226
|
KIAA0226 |
| chr16_+_29984962 | 0.14 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr19_+_50084561 | 0.14 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr17_+_39969183 | 0.14 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr19_-_46272106 | 0.14 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr17_-_76124812 | 0.14 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr11_+_46402583 | 0.14 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr16_-_2318055 | 0.14 |
ENST00000561518.1
ENST00000561718.1 ENST00000567147.1 ENST00000562690.1 ENST00000569598.2 |
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr10_-_49813090 | 0.14 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr9_+_131218698 | 0.14 |
ENST00000434106.3
ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr10_+_99400443 | 0.14 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr11_+_706219 | 0.14 |
ENST00000533500.1
|
EPS8L2
|
EPS8-like 2 |
| chr14_-_24711806 | 0.14 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_-_10415218 | 0.14 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr9_+_140149625 | 0.14 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr19_-_11039261 | 0.14 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr7_+_101917407 | 0.14 |
ENST00000487284.1
|
CUX1
|
cut-like homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.5 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.7 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.3 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.1 | 0.4 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.3 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.3 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.4 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.4 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.3 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.4 | GO:0030578 | PML body organization(GO:0030578) MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.2 | GO:1902683 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.5 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of anagen(GO:0051885) |
| 0.0 | 0.4 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.0 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:0032639 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.2 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0021557 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0010935 | regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.5 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0036435 | IkappaB kinase activity(GO:0008384) K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |