Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
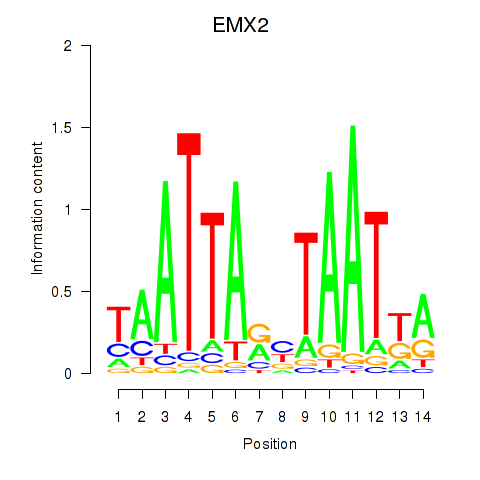
Results for EMX2
Z-value: 1.04
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.10 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg19_v2_chr10_+_119301928_119301955 | -0.26 | 6.2e-01 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_174818390 | 1.50 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr15_+_63188009 | 0.85 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr14_+_20187174 | 0.76 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr8_-_40200903 | 0.75 |
ENST00000522486.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr11_+_327171 | 0.71 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr2_+_172309634 | 0.63 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr1_+_159931002 | 0.54 |
ENST00000443364.1
ENST00000423943.1 |
RP11-48O20.4
|
long intergenic non-protein coding RNA 1133 |
| chr11_+_12308447 | 0.53 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr19_+_23257988 | 0.48 |
ENST00000594318.1
ENST00000600223.1 ENST00000593635.1 |
CTD-2291D10.4
ZNF730
|
CTD-2291D10.4 zinc finger protein 730 |
| chr11_-_58378759 | 0.47 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr4_+_80584903 | 0.45 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr2_-_99279928 | 0.45 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr6_+_13272904 | 0.43 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr3_-_12587055 | 0.42 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr6_-_26235206 | 0.40 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr21_+_35553045 | 0.40 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr8_-_6115044 | 0.37 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr11_-_63376013 | 0.36 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr16_+_15489629 | 0.35 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr7_-_17598506 | 0.35 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr12_+_21207503 | 0.35 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr1_+_56046710 | 0.35 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr4_+_76871883 | 0.34 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr12_-_51418549 | 0.34 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr5_+_136070614 | 0.34 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr8_-_133687813 | 0.31 |
ENST00000250173.1
ENST00000519595.1 |
LRRC6
|
leucine rich repeat containing 6 |
| chr22_-_40929812 | 0.31 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr5_+_32788945 | 0.30 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr3_-_155011483 | 0.30 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr11_+_45376922 | 0.30 |
ENST00000524410.1
ENST00000524488.1 ENST00000524565.1 |
RP11-430H10.1
|
RP11-430H10.1 |
| chr1_-_67142710 | 0.29 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr11_-_104905840 | 0.29 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr22_+_40440804 | 0.28 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr19_+_54466179 | 0.28 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr16_+_15489603 | 0.28 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr6_+_63921399 | 0.27 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr12_+_25055243 | 0.27 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr17_+_61151306 | 0.27 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr15_-_34447023 | 0.27 |
ENST00000560310.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr21_-_19858196 | 0.27 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chrM_+_10758 | 0.26 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr10_-_21186144 | 0.26 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr22_+_50981079 | 0.26 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr9_-_99540328 | 0.25 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr12_+_72061563 | 0.25 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr12_+_1099675 | 0.25 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chrX_+_100224676 | 0.24 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr16_-_74330612 | 0.24 |
ENST00000569389.1
ENST00000562888.1 |
AC009120.4
|
AC009120.4 |
| chr4_+_95128748 | 0.23 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr8_-_133687778 | 0.23 |
ENST00000518642.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr12_-_10007448 | 0.22 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chrX_-_9734004 | 0.22 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr2_+_161993465 | 0.22 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr3_-_130745403 | 0.22 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr10_-_99771079 | 0.21 |
ENST00000309155.3
|
CRTAC1
|
cartilage acidic protein 1 |
| chr2_-_14541060 | 0.21 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr1_+_178482262 | 0.20 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr8_+_107593198 | 0.20 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr4_-_71532207 | 0.20 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr11_-_104916034 | 0.18 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr17_+_7239821 | 0.18 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr15_+_75970672 | 0.18 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr2_-_231989808 | 0.18 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_56212270 | 0.18 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr11_+_119019722 | 0.18 |
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr3_-_194188956 | 0.18 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chrX_+_77166172 | 0.17 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr15_-_81202118 | 0.17 |
ENST00000560560.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr4_-_89205879 | 0.17 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr8_+_31497271 | 0.17 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr4_+_156824840 | 0.17 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr15_-_57025759 | 0.17 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr16_+_22524844 | 0.17 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr10_-_90611566 | 0.17 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr19_-_35264089 | 0.16 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr1_+_78470530 | 0.16 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr12_-_74686314 | 0.16 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr1_-_119682812 | 0.16 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chrX_+_47444613 | 0.16 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr3_+_119298523 | 0.15 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr5_-_111754948 | 0.15 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr14_+_85994943 | 0.15 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr15_-_99057551 | 0.14 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr12_-_8803128 | 0.14 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_86650077 | 0.14 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_+_63921351 | 0.14 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr4_-_103746924 | 0.14 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_179343226 | 0.14 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chrX_-_72434628 | 0.14 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr17_-_39093672 | 0.14 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr11_+_125439176 | 0.14 |
ENST00000529812.1
|
EI24
|
etoposide induced 2.4 |
| chr16_-_30102547 | 0.13 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr4_-_103747011 | 0.13 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_+_77713222 | 0.13 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr8_+_119294456 | 0.13 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr12_+_59989918 | 0.13 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr4_-_109541610 | 0.13 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr5_-_111312622 | 0.13 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr4_+_158493642 | 0.13 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr5_-_74162739 | 0.13 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr10_-_79397740 | 0.12 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_186692745 | 0.12 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr2_-_198540751 | 0.12 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr2_+_67624430 | 0.12 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr17_+_79849872 | 0.12 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr4_+_56212505 | 0.12 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr5_-_111092930 | 0.12 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr6_+_26204825 | 0.12 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr6_+_153552455 | 0.12 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr17_+_67590125 | 0.12 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr16_-_21868978 | 0.12 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr10_-_115904361 | 0.12 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr19_-_33360647 | 0.12 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr11_+_66276550 | 0.11 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr5_+_179135246 | 0.11 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr9_-_47314222 | 0.11 |
ENST00000420228.1
ENST00000438517.1 ENST00000414020.1 |
AL953854.2
|
AL953854.2 |
| chr12_+_94071129 | 0.11 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr8_+_110552337 | 0.11 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr12_+_29302119 | 0.11 |
ENST00000536681.3
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr2_-_55237484 | 0.11 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr12_-_100656134 | 0.11 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr1_-_170043709 | 0.11 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr16_-_29415350 | 0.11 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr2_+_11674213 | 0.11 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr14_+_95027772 | 0.11 |
ENST00000555095.1
ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr2_+_196313239 | 0.11 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr9_+_131902346 | 0.11 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr4_-_185275104 | 0.10 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr2_+_183982255 | 0.10 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr8_-_123139423 | 0.10 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr4_+_71384300 | 0.10 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr12_-_112279694 | 0.10 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr5_-_76788317 | 0.10 |
ENST00000296679.4
|
WDR41
|
WD repeat domain 41 |
| chr16_+_69599861 | 0.10 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr12_-_71533055 | 0.10 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr19_-_44388116 | 0.10 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr16_-_28937027 | 0.10 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr20_+_9494987 | 0.10 |
ENST00000427562.2
ENST00000246070.2 |
LAMP5
|
lysosomal-associated membrane protein family, member 5 |
| chr2_-_44550441 | 0.10 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr2_+_44001172 | 0.10 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr14_-_60043524 | 0.10 |
ENST00000537690.2
ENST00000281581.4 |
CCDC175
|
coiled-coil domain containing 175 |
| chr11_+_122753391 | 0.10 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr14_+_52164820 | 0.09 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr4_-_68620053 | 0.09 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr12_+_64798095 | 0.09 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr9_-_104198042 | 0.09 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr5_-_157161727 | 0.09 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr1_+_146373546 | 0.09 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr1_-_35450897 | 0.08 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr2_+_11696464 | 0.08 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr7_+_138145076 | 0.08 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr4_-_39034542 | 0.08 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr12_+_94071341 | 0.08 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr2_+_120687335 | 0.08 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chrX_-_23926004 | 0.08 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr6_+_34725181 | 0.08 |
ENST00000244520.5
|
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr1_-_39347255 | 0.07 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr8_-_13134246 | 0.07 |
ENST00000503161.2
|
DLC1
|
deleted in liver cancer 1 |
| chr4_-_109541539 | 0.07 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr11_-_4719072 | 0.07 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chrX_-_72347916 | 0.07 |
ENST00000373518.1
|
NAP1L6
|
nucleosome assembly protein 1-like 6 |
| chr3_-_186288097 | 0.07 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr3_+_63638372 | 0.07 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr16_+_29832634 | 0.07 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr5_+_111755280 | 0.07 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr3_+_12598563 | 0.07 |
ENST00000411987.1
ENST00000448482.1 |
MKRN2
|
makorin ring finger protein 2 |
| chr13_-_108867101 | 0.07 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr4_+_89206076 | 0.07 |
ENST00000500009.2
|
RP11-10L7.1
|
RP11-10L7.1 |
| chr14_+_62164340 | 0.07 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr2_+_234826016 | 0.07 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr5_-_130970723 | 0.07 |
ENST00000308008.6
ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr8_+_35649365 | 0.07 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr5_-_137475071 | 0.07 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr19_-_39340563 | 0.06 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr1_-_54405773 | 0.06 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr5_-_140013275 | 0.06 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr11_+_74303575 | 0.06 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr1_-_85870177 | 0.06 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr9_+_71986182 | 0.06 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr15_-_101137121 | 0.06 |
ENST00000560934.1
|
LINS
|
lines homolog (Drosophila) |
| chr5_+_112312416 | 0.06 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr1_+_174844645 | 0.06 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_-_10282742 | 0.06 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr14_+_55494323 | 0.06 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr21_+_35552978 | 0.06 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr8_-_57123815 | 0.06 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr14_+_55493920 | 0.06 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr5_-_76788298 | 0.06 |
ENST00000511036.1
|
WDR41
|
WD repeat domain 41 |
| chr1_+_81771806 | 0.06 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chrX_-_15402498 | 0.06 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr1_+_225600404 | 0.06 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr7_+_129906660 | 0.06 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr11_-_102714534 | 0.06 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr19_-_37697976 | 0.06 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
| chr6_-_109761707 | 0.06 |
ENST00000520723.1
ENST00000518648.1 ENST00000417394.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr11_-_107729287 | 0.06 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_170897045 | 0.06 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr4_+_25162253 | 0.06 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr16_-_1538765 | 0.06 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chrX_+_55744228 | 0.06 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |