Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
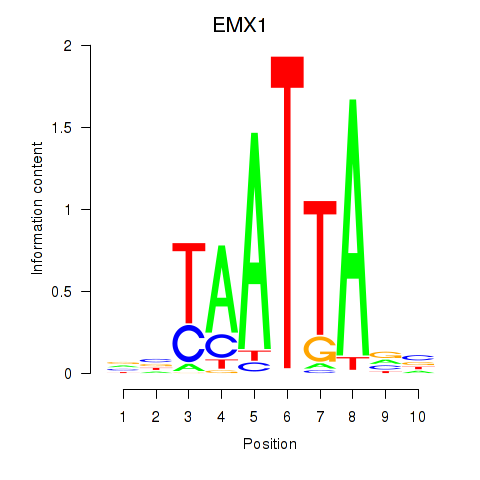
Results for EMX1
Z-value: 0.36
Transcription factors associated with EMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX1
|
ENSG00000135638.9 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX1 | hg19_v2_chr2_+_73144604_73144654 | -0.01 | 9.8e-01 | Click! |
Activity profile of EMX1 motif
Sorted Z-values of EMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_110958124 | 0.34 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr8_-_1922789 | 0.30 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr17_+_19091325 | 0.27 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr15_-_53002007 | 0.26 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
| chr15_+_58702742 | 0.22 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr14_+_97925151 | 0.19 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr8_+_94752349 | 0.18 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr2_-_99871570 | 0.18 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr1_-_183538319 | 0.18 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_+_202317855 | 0.18 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr8_+_99956662 | 0.17 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr19_+_48949087 | 0.17 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr4_-_89442940 | 0.16 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr14_+_100485712 | 0.16 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr14_+_39944025 | 0.15 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr3_-_12587055 | 0.15 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chrX_+_10031499 | 0.14 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr4_+_159727272 | 0.14 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr16_+_32264645 | 0.14 |
ENST00000569631.1
ENST00000354614.3 |
TP53TG3D
|
TP53 target 3D |
| chr17_-_19015945 | 0.14 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr17_-_39150385 | 0.14 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr17_+_61151306 | 0.14 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr7_-_99716914 | 0.14 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_-_21625486 | 0.14 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr2_-_27498208 | 0.14 |
ENST00000424577.1
ENST00000426569.1 |
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr11_+_72983246 | 0.14 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr14_+_93389425 | 0.13 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr13_-_30880979 | 0.13 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr6_+_76599809 | 0.13 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr4_+_129732419 | 0.13 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr6_+_148593425 | 0.13 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr13_-_30160925 | 0.13 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_+_107712173 | 0.12 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr8_-_42396185 | 0.12 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr8_+_77593448 | 0.12 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_+_42187608 | 0.12 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr20_+_60174827 | 0.12 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr4_-_122085469 | 0.11 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr7_+_129984630 | 0.11 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr9_+_131037623 | 0.11 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr12_-_25348007 | 0.11 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr6_+_26365443 | 0.11 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr14_-_70883708 | 0.11 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr8_+_99956759 | 0.11 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr1_-_234667504 | 0.11 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr19_-_46088068 | 0.11 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr11_-_111794446 | 0.11 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr17_-_33446735 | 0.11 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr15_+_62853562 | 0.11 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr17_-_55911970 | 0.11 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr3_+_111393501 | 0.11 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr3_+_156799587 | 0.11 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr1_+_10509971 | 0.11 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr7_+_99717230 | 0.11 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr7_+_23719749 | 0.10 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr12_-_46121554 | 0.10 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr2_+_27498289 | 0.10 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr19_+_48949030 | 0.10 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr15_-_72565340 | 0.10 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr6_-_26032288 | 0.10 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr17_-_46799872 | 0.10 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr2_-_214016314 | 0.10 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_+_202317815 | 0.10 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_-_117747327 | 0.10 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr9_-_75488984 | 0.10 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr22_-_33968239 | 0.10 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr7_-_14029283 | 0.10 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr20_+_42187682 | 0.10 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_9953295 | 0.10 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr1_+_154401791 | 0.10 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr2_-_1629176 | 0.10 |
ENST00000366424.2
|
AC144450.2
|
AC144450.2 |
| chr11_-_111649074 | 0.10 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_+_131438443 | 0.09 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr12_+_26348246 | 0.09 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr2_-_73460334 | 0.09 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr4_+_169418255 | 0.09 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr16_+_69345243 | 0.09 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr2_+_28618532 | 0.09 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr6_+_13272904 | 0.09 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr8_+_98900132 | 0.09 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_+_150065278 | 0.09 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr14_+_85994943 | 0.09 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr17_+_73455788 | 0.09 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr2_+_166095898 | 0.09 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_-_26235206 | 0.09 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr6_-_33256664 | 0.09 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr16_+_12059091 | 0.09 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr9_-_139965000 | 0.09 |
ENST00000409687.3
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr11_-_26593649 | 0.08 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chrX_+_135730297 | 0.08 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr10_-_75351088 | 0.08 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr17_-_39623681 | 0.08 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr13_-_47471155 | 0.08 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr7_+_79998864 | 0.08 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr21_-_43346790 | 0.08 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr2_+_7073174 | 0.08 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr2_-_203735586 | 0.08 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_-_57328187 | 0.08 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr14_+_32798547 | 0.08 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr14_-_95236551 | 0.08 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr7_-_86849836 | 0.08 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr16_+_14280742 | 0.08 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_-_159832438 | 0.08 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr16_-_4852915 | 0.08 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr6_+_26199737 | 0.08 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chrX_+_107288197 | 0.08 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_+_187957646 | 0.08 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_-_100860851 | 0.08 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr22_+_44427230 | 0.08 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr15_+_59910132 | 0.08 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr17_-_56296580 | 0.08 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr2_-_216003127 | 0.08 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr17_-_46690839 | 0.08 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr4_+_169418195 | 0.07 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_+_13671225 | 0.07 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr4_+_3388057 | 0.07 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr1_-_232651312 | 0.07 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_-_201140673 | 0.07 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr21_+_33671160 | 0.07 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr15_-_72563585 | 0.07 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr2_+_158114051 | 0.07 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr17_-_73663245 | 0.07 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr2_-_197226875 | 0.07 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr19_-_56110859 | 0.07 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr10_+_17270214 | 0.07 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr1_+_222910625 | 0.07 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr16_+_53412368 | 0.07 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr12_+_48592134 | 0.07 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr9_-_107690420 | 0.07 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr7_-_99716952 | 0.07 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_-_74486347 | 0.07 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_-_130745403 | 0.07 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr8_-_124553437 | 0.07 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr5_+_66300464 | 0.07 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_7014064 | 0.07 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr6_-_139613269 | 0.07 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr19_+_18942720 | 0.07 |
ENST00000262803.5
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr12_-_42878101 | 0.07 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr8_-_116504448 | 0.07 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr19_-_14064114 | 0.07 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr17_-_1532106 | 0.07 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr12_-_56321397 | 0.07 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr5_+_137722255 | 0.07 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr18_+_46065393 | 0.07 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr17_-_40288449 | 0.07 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr6_-_127840336 | 0.07 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr12_+_26348429 | 0.07 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chrX_-_15332665 | 0.07 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr19_+_18942761 | 0.07 |
ENST00000599848.1
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr6_-_138866823 | 0.07 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr10_-_116418053 | 0.07 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr16_+_28505955 | 0.07 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
| chr12_-_56693758 | 0.07 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr15_-_99789736 | 0.07 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr7_+_100860949 | 0.07 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr17_-_72772462 | 0.06 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr17_-_38928414 | 0.06 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr14_-_73360796 | 0.06 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr17_-_28661065 | 0.06 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr17_+_42785976 | 0.06 |
ENST00000393547.2
ENST00000398338.3 |
DBF4B
|
DBF4 homolog B (S. cerevisiae) |
| chr4_-_19458597 | 0.06 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr3_+_152552685 | 0.06 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr17_-_74163159 | 0.06 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr12_+_7013897 | 0.06 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr8_+_77593474 | 0.06 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr7_-_99717463 | 0.06 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_111032971 | 0.06 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_+_58430567 | 0.06 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr3_-_193096600 | 0.06 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr7_+_55433131 | 0.06 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr20_+_33292507 | 0.06 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr11_-_102709441 | 0.06 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr19_-_39368887 | 0.06 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chrX_+_149887090 | 0.06 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr4_-_52883786 | 0.06 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr14_+_74034310 | 0.06 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr16_+_103816 | 0.06 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr18_+_59415396 | 0.06 |
ENST00000567801.1
|
RP11-1096D5.1
|
RP11-1096D5.1 |
| chr6_+_161123270 | 0.06 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr13_-_38172863 | 0.06 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr7_-_99716940 | 0.06 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr20_+_30697298 | 0.06 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr12_-_3862245 | 0.06 |
ENST00000252322.1
ENST00000440314.2 |
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr17_-_19364269 | 0.06 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr9_+_12693336 | 0.06 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr10_+_80027105 | 0.06 |
ENST00000461034.1
ENST00000476909.1 ENST00000459633.1 |
LINC00595
|
long intergenic non-protein coding RNA 595 |
| chr12_-_11091862 | 0.06 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr7_+_99425633 | 0.06 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr2_-_190446738 | 0.06 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr16_+_14280564 | 0.06 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr2_-_74619152 | 0.06 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr16_+_58010339 | 0.06 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr22_+_30752606 | 0.06 |
ENST00000399824.2
ENST00000405659.1 ENST00000338306.3 |
CCDC157
|
coiled-coil domain containing 157 |
| chr10_-_5046042 | 0.06 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr1_-_94586651 | 0.06 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr2_+_109237717 | 0.06 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_+_58430368 | 0.06 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chrX_+_78003204 | 0.05 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr3_-_151176497 | 0.05 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr13_-_74993252 | 0.05 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.3 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0060345 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.0 | GO:0060928 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 0.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0015254 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |