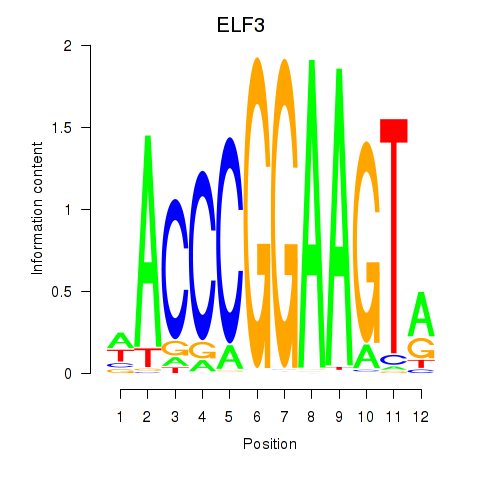
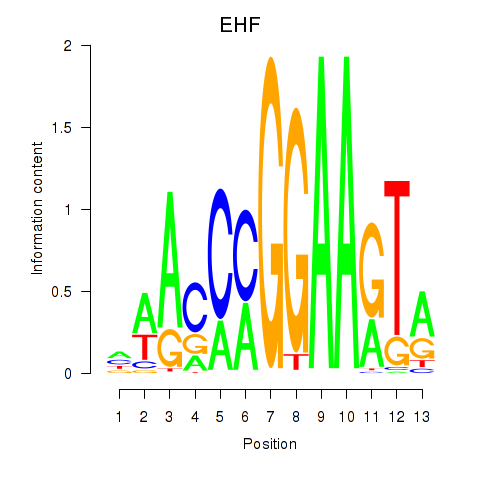
| 0.4 |
1.3 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.3 |
0.9 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 |
1.0 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.2 |
0.7 |
GO:0000349 |
generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.2 |
0.2 |
GO:0002590 |
regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.2 |
0.8 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 |
0.6 |
GO:0016334 |
morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 |
1.3 |
GO:1904327 |
protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 |
0.8 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.1 |
0.7 |
GO:0061624 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 |
0.3 |
GO:1903450 |
regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 |
0.5 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 |
0.3 |
GO:1900738 |
dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 |
0.3 |
GO:0032581 |
ER-dependent peroxisome organization(GO:0032581) |
| 0.1 |
0.4 |
GO:0018282 |
metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 |
0.7 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 |
0.4 |
GO:0071348 |
cellular response to interleukin-11(GO:0071348) |
| 0.1 |
0.6 |
GO:0071486 |
cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 |
0.3 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
0.3 |
GO:1902598 |
creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.1 |
0.3 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.1 |
0.5 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.4 |
GO:0015783 |
GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 |
0.2 |
GO:1904976 |
response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 |
0.5 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.4 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.4 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
1.3 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.2 |
GO:0048213 |
Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 |
0.6 |
GO:0017185 |
peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 |
0.1 |
GO:0051081 |
mitotic nuclear envelope disassembly(GO:0007077) membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 |
0.2 |
GO:0032747 |
positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 |
0.2 |
GO:0002725 |
negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 |
0.2 |
GO:0060086 |
circadian temperature homeostasis(GO:0060086) |
| 0.1 |
0.1 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 |
0.7 |
GO:1903944 |
regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 |
0.3 |
GO:0019086 |
late viral transcription(GO:0019086) |
| 0.1 |
0.2 |
GO:1990502 |
dense core granule maturation(GO:1990502) |
| 0.1 |
0.3 |
GO:1904751 |
positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 |
0.2 |
GO:0001300 |
chronological cell aging(GO:0001300) |
| 0.1 |
0.8 |
GO:1990416 |
cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 |
0.2 |
GO:0055099 |
response to high density lipoprotein particle(GO:0055099) |
| 0.1 |
0.2 |
GO:0042377 |
menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 |
0.3 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
0.2 |
GO:1902824 |
cleavage furrow ingression(GO:0036090) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 |
0.8 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.4 |
GO:0051715 |
cytolysis in other organism(GO:0051715) |
| 0.1 |
0.1 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 |
0.1 |
GO:0019520 |
aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 |
0.9 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.2 |
GO:0090579 |
transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 |
0.1 |
GO:0006425 |
glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 |
0.1 |
GO:1902463 |
protein localization to cell leading edge(GO:1902463) |
| 0.0 |
1.3 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.9 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 |
0.1 |
GO:0045590 |
negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 |
0.3 |
GO:0033031 |
positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 |
0.1 |
GO:2000360 |
negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 |
0.7 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.3 |
GO:0043126 |
regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 |
0.1 |
GO:2000619 |
negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 |
0.3 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.7 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.2 |
GO:0043376 |
regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 |
0.2 |
GO:0039689 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 |
0.5 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.3 |
GO:0010836 |
negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 |
0.7 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.2 |
GO:1905224 |
clathrin-coated pit assembly(GO:1905224) |
| 0.0 |
0.2 |
GO:0036343 |
psychomotor behavior(GO:0036343) |
| 0.0 |
0.1 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 |
0.4 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.0 |
0.1 |
GO:0035674 |
tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 |
0.2 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 |
0.0 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.0 |
0.2 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 |
0.3 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 |
0.1 |
GO:0042737 |
drug catabolic process(GO:0042737) |
| 0.0 |
0.3 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.2 |
GO:0036496 |
regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 |
0.1 |
GO:0043309 |
regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 |
0.3 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 |
0.1 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 |
0.3 |
GO:0046618 |
drug export(GO:0046618) |
| 0.0 |
0.5 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.2 |
GO:0019303 |
D-ribose catabolic process(GO:0019303) |
| 0.0 |
0.2 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.0 |
0.2 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 |
0.1 |
GO:0048210 |
Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 |
0.3 |
GO:0017182 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.2 |
GO:0090467 |
L-arginine import(GO:0043091) divalent metal ion export(GO:0070839) arginine import(GO:0090467) |
| 0.0 |
0.1 |
GO:0048597 |
B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 |
0.1 |
GO:0070902 |
mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 |
0.2 |
GO:1904925 |
negative regulation of mitochondrial fusion(GO:0010637) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 |
0.2 |
GO:0046985 |
positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 |
0.2 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
0.1 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 |
0.3 |
GO:2000622 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.1 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 |
0.4 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.0 |
0.3 |
GO:0009146 |
purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 |
0.0 |
GO:1904640 |
response to methionine(GO:1904640) |
| 0.0 |
0.1 |
GO:0010897 |
negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 |
0.1 |
GO:0035054 |
embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 |
0.3 |
GO:2000002 |
negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 |
0.2 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.0 |
0.4 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.2 |
GO:0071440 |
regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.1 |
GO:0038155 |
interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 |
0.1 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 |
0.1 |
GO:0048320 |
axial mesoderm formation(GO:0048320) |
| 0.0 |
0.6 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.6 |
GO:0060044 |
negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 |
0.4 |
GO:0035751 |
regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 |
0.6 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.1 |
GO:0042727 |
flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 |
0.1 |
GO:0046901 |
tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 |
0.1 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.0 |
0.1 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 |
0.2 |
GO:2000784 |
regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 |
0.2 |
GO:1903385 |
regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 |
0.1 |
GO:0002276 |
basophil activation involved in immune response(GO:0002276) |
| 0.0 |
0.1 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 |
0.1 |
GO:0002399 |
MHC class II protein complex assembly(GO:0002399) |
| 0.0 |
0.1 |
GO:0033319 |
UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.0 |
0.2 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.0 |
0.2 |
GO:0002084 |
protein depalmitoylation(GO:0002084) |
| 0.0 |
0.2 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
0.2 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.0 |
0.1 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.0 |
0.2 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 |
0.1 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 |
0.1 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.0 |
0.3 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.0 |
0.2 |
GO:0031087 |
deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.1 |
GO:1904222 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.1 |
GO:0061737 |
leukotriene signaling pathway(GO:0061737) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.4 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.2 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.1 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.0 |
0.1 |
GO:0071847 |
TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 |
0.1 |
GO:0052551 |
response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 |
0.3 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.1 |
GO:0003292 |
cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 |
0.8 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.0 |
0.0 |
GO:1990167 |
protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 |
0.4 |
GO:0031163 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:1904526 |
regulation of microtubule binding(GO:1904526) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.1 |
GO:0043012 |
regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 |
0.3 |
GO:0071688 |
striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 |
0.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.2 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 |
0.3 |
GO:0097012 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 |
0.0 |
GO:1901979 |
regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 |
0.1 |
GO:2000537 |
regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 |
0.1 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.0 |
0.1 |
GO:0035825 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 |
0.2 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.4 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 |
0.3 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.0 |
GO:0010980 |
regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) positive regulation of response to alcohol(GO:1901421) |
| 0.0 |
0.5 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
0.3 |
GO:0030207 |
chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 |
0.2 |
GO:0034384 |
high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 |
0.0 |
GO:1903336 |
negative regulation of vacuolar transport(GO:1903336) |
| 0.0 |
0.3 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.3 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.1 |
GO:0002729 |
positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 |
0.1 |
GO:0000270 |
peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 |
0.3 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.1 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 |
0.2 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 |
0.2 |
GO:2001300 |
lipoxin metabolic process(GO:2001300) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:0097398 |
response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 |
0.1 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
0.0 |
GO:0002906 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 |
0.0 |
GO:0061713 |
neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 |
0.2 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.0 |
GO:0036493 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 |
0.1 |
GO:0035026 |
leading edge cell differentiation(GO:0035026) |
| 0.0 |
0.0 |
GO:1901656 |
glycoside transport(GO:1901656) |
| 0.0 |
0.1 |
GO:0002326 |
B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 |
0.1 |
GO:0035720 |
intraciliary anterograde transport(GO:0035720) |
| 0.0 |
0.1 |
GO:0000466 |
maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 |
0.1 |
GO:0051304 |
chromosome separation(GO:0051304) |
| 0.0 |
0.1 |
GO:0061767 |
negative regulation of lung blood pressure(GO:0061767) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.2 |
GO:0014029 |
neural crest formation(GO:0014029) |
| 0.0 |
0.1 |
GO:0090410 |
malonate catabolic process(GO:0090410) |
| 0.0 |
0.0 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.1 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 |
0.1 |
GO:1901073 |
N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 |
0.6 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.0 |
0.0 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.0 |
0.1 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 |
0.1 |
GO:0060546 |
negative regulation of necroptotic process(GO:0060546) protein linear polyubiquitination(GO:0097039) |
| 0.0 |
0.1 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.2 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 |
0.1 |
GO:1902846 |
regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 |
0.1 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.1 |
GO:0055129 |
L-proline biosynthetic process(GO:0055129) |
| 0.0 |
0.1 |
GO:0070200 |
establishment of protein localization to telomere(GO:0070200) |
| 0.0 |
0.1 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
0.0 |
GO:0002583 |
regulation of antigen processing and presentation of peptide antigen(GO:0002583) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 |
0.0 |
GO:0019516 |
lactate oxidation(GO:0019516) |
| 0.0 |
0.0 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.0 |
0.1 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.0 |
GO:0035865 |
cellular response to potassium ion(GO:0035865) |
| 0.0 |
0.1 |
GO:0002191 |
cap-dependent translational initiation(GO:0002191) |
| 0.0 |
0.0 |
GO:0072334 |
UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 |
0.1 |
GO:0072752 |
cellular response to rapamycin(GO:0072752) |
| 0.0 |
0.0 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 |
0.1 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.7 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.0 |
0.0 |
GO:0042823 |
pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 |
0.0 |
GO:0072361 |
regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 |
0.4 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.0 |
0.0 |
GO:0045083 |
negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.0 |
GO:0043328 |
protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.4 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
0.4 |
GO:1901186 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
0.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.0 |
GO:0098976 |
excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 |
0.4 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.7 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:0033088 |
regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 |
0.2 |
GO:0048505 |
regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 |
0.2 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 |
0.0 |
GO:0046968 |
peptide antigen transport(GO:0046968) |
| 0.0 |
0.1 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.1 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 |
0.1 |
GO:0070129 |
regulation of mitochondrial translation(GO:0070129) |
| 0.0 |
0.3 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.3 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.2 |
GO:0050995 |
negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 |
0.1 |
GO:0098967 |
exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 |
0.1 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.0 |
0.4 |
GO:0031063 |
regulation of histone deacetylation(GO:0031063) |
| 0.0 |
0.1 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.1 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |