Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
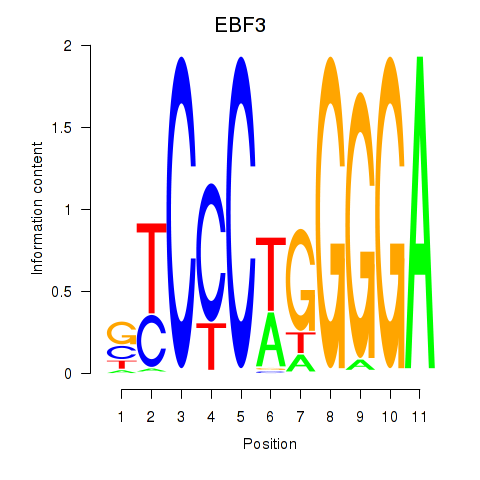
Results for EBF3
Z-value: 1.62
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.9 | EBF transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF3 | hg19_v2_chr10_-_131640435_131640435 | 0.23 | 6.6e-01 | Click! |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_38065052 | 1.02 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr7_-_100881041 | 0.96 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr2_+_238499812 | 0.92 |
ENST00000452801.1
|
AC104667.3
|
AC104667.3 |
| chr1_-_41950342 | 0.88 |
ENST00000372587.4
|
EDN2
|
endothelin 2 |
| chr11_+_20044375 | 0.88 |
ENST00000525322.1
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr15_-_40633101 | 0.88 |
ENST00000559313.1
|
C15orf52
|
chromosome 15 open reading frame 52 |
| chr1_+_23695680 | 0.80 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr19_+_45909893 | 0.76 |
ENST00000592852.1
ENST00000589804.1 ENST00000590794.1 |
CD3EAP
|
CD3e molecule, epsilon associated protein |
| chr17_+_27052892 | 0.75 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr12_-_106697974 | 0.74 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr17_-_7832753 | 0.71 |
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr11_+_118978045 | 0.70 |
ENST00000336702.3
|
C2CD2L
|
C2CD2-like |
| chr7_-_155604967 | 0.67 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr11_+_20044096 | 0.66 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr17_+_75315654 | 0.64 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr11_-_9482010 | 0.63 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr10_+_73724123 | 0.63 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr5_-_175964366 | 0.61 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr17_-_42994283 | 0.60 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr6_+_292253 | 0.59 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr16_+_19619083 | 0.59 |
ENST00000538552.1
|
C16orf62
|
chromosome 16 open reading frame 62 |
| chr4_-_90758227 | 0.58 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr16_+_30662050 | 0.58 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr16_+_3068393 | 0.57 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr11_+_124735282 | 0.56 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr16_+_70207686 | 0.56 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr15_+_66994561 | 0.56 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr6_+_148593425 | 0.55 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr6_-_30654977 | 0.55 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr14_-_102704783 | 0.55 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chr19_+_55996565 | 0.53 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr16_+_30662085 | 0.53 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr11_-_69294647 | 0.53 |
ENST00000542064.1
|
AP000439.3
|
AP000439.3 |
| chr14_+_24590560 | 0.53 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr19_-_40772221 | 0.52 |
ENST00000441941.2
ENST00000580747.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_+_73629500 | 0.52 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr16_+_81772633 | 0.51 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chrX_+_9217932 | 0.50 |
ENST00000432442.1
|
GS1-519E5.1
|
GS1-519E5.1 |
| chrX_-_49056635 | 0.50 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr17_+_75315534 | 0.50 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr17_-_1395954 | 0.48 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr1_+_33231221 | 0.47 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522 |
| chr1_-_21995794 | 0.47 |
ENST00000542643.2
ENST00000374765.4 ENST00000317967.7 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr2_+_27301435 | 0.47 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr18_-_72920372 | 0.47 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr22_-_19466643 | 0.47 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr19_-_1650666 | 0.47 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr19_+_984313 | 0.46 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chrX_+_47083037 | 0.46 |
ENST00000523034.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr7_+_26438187 | 0.46 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr5_+_159656437 | 0.46 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr1_+_110577229 | 0.46 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr3_+_159481464 | 0.46 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr7_-_100287071 | 0.46 |
ENST00000275732.5
|
GIGYF1
|
GRB10 interacting GYF protein 1 |
| chr16_-_29478016 | 0.46 |
ENST00000549858.1
ENST00000551411.1 |
RP11-345J4.3
|
Uncharacterized protein |
| chr16_+_67233007 | 0.45 |
ENST00000360833.1
ENST00000393997.2 |
ELMO3
|
engulfment and cell motility 3 |
| chr11_+_20044600 | 0.45 |
ENST00000311043.8
|
NAV2
|
neuron navigator 2 |
| chr10_+_77056181 | 0.45 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr1_-_24438664 | 0.45 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chr1_+_110453462 | 0.44 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_110453203 | 0.44 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr22_+_45705987 | 0.44 |
ENST00000405673.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr22_+_29999545 | 0.43 |
ENST00000413209.2
ENST00000347330.5 ENST00000338641.4 ENST00000403435.1 ENST00000361452.4 ENST00000403999.3 |
NF2
|
neurofibromin 2 (merlin) |
| chr19_-_4722705 | 0.43 |
ENST00000598360.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr6_+_37012607 | 0.43 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr3_+_184018352 | 0.42 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chrX_-_153637612 | 0.42 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr19_+_17638059 | 0.42 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr22_+_18560743 | 0.42 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr20_+_62496596 | 0.42 |
ENST00000369927.4
ENST00000346249.4 ENST00000348257.5 ENST00000352482.4 ENST00000351424.4 ENST00000217121.5 ENST00000358548.4 |
TPD52L2
|
tumor protein D52-like 2 |
| chr17_+_73452545 | 0.42 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr16_+_69958887 | 0.41 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr14_-_89960395 | 0.41 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr16_-_67867749 | 0.41 |
ENST00000566758.1
ENST00000445712.2 ENST00000219172.3 ENST00000564817.1 |
CENPT
|
centromere protein T |
| chr12_-_291556 | 0.41 |
ENST00000537295.1
ENST00000537961.1 |
RP11-598F7.6
|
RP11-598F7.6 |
| chr11_-_62446527 | 0.41 |
ENST00000294119.2
ENST00000529640.1 ENST00000534176.1 ENST00000301935.5 |
UBXN1
|
UBX domain protein 1 |
| chr1_+_110453514 | 0.41 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr17_-_80023401 | 0.40 |
ENST00000354321.7
ENST00000306796.5 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr4_+_4861385 | 0.40 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr6_+_42984723 | 0.40 |
ENST00000332245.8
|
KLHDC3
|
kelch domain containing 3 |
| chr11_+_22646739 | 0.40 |
ENST00000428556.2
|
AC103801.2
|
AC103801.2 |
| chr5_-_146833222 | 0.40 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr6_+_43738444 | 0.40 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr11_-_61124776 | 0.39 |
ENST00000542361.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr17_-_42295870 | 0.39 |
ENST00000526094.1
ENST00000529383.1 ENST00000530828.1 |
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr19_+_35634146 | 0.39 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr2_-_31361543 | 0.39 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr7_-_100881109 | 0.39 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr7_+_100464760 | 0.39 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr5_+_139028510 | 0.38 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr16_+_4896659 | 0.38 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr7_+_29874341 | 0.38 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr16_+_30662184 | 0.37 |
ENST00000300835.4
|
PRR14
|
proline rich 14 |
| chr3_+_193853927 | 0.37 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr17_-_79881408 | 0.37 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr22_-_23922410 | 0.36 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr15_-_83240783 | 0.36 |
ENST00000568994.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr22_+_29702572 | 0.36 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr16_-_30798492 | 0.36 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr14_-_106994333 | 0.36 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr17_-_7493390 | 0.35 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr1_-_154943212 | 0.35 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr1_+_156589198 | 0.35 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr22_+_21321447 | 0.35 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr20_+_33134619 | 0.35 |
ENST00000374837.3
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr7_-_27187393 | 0.35 |
ENST00000222728.3
|
HOXA6
|
homeobox A6 |
| chr17_-_42296855 | 0.35 |
ENST00000436088.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr1_-_26197744 | 0.34 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr14_-_23540826 | 0.34 |
ENST00000357481.2
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr15_-_86338134 | 0.34 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr5_-_16738451 | 0.34 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr17_-_43045439 | 0.34 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr17_+_73539232 | 0.34 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr7_+_110731062 | 0.34 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr1_-_27693349 | 0.34 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr7_+_100183927 | 0.34 |
ENST00000241071.6
ENST00000360609.2 |
FBXO24
|
F-box protein 24 |
| chr16_+_30662360 | 0.34 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr19_+_50433476 | 0.34 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr2_+_206950095 | 0.34 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr17_-_80023659 | 0.33 |
ENST00000578907.1
ENST00000577907.1 ENST00000578176.1 ENST00000582529.1 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr10_-_47151341 | 0.33 |
ENST00000422732.2
|
LINC00842
|
long intergenic non-protein coding RNA 842 |
| chr16_-_68482440 | 0.33 |
ENST00000219334.5
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr1_-_156460391 | 0.33 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr1_-_120190396 | 0.33 |
ENST00000421812.2
|
ZNF697
|
zinc finger protein 697 |
| chr20_-_30795511 | 0.33 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr17_-_42580738 | 0.33 |
ENST00000585614.1
ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8
|
G patch domain containing 8 |
| chr9_+_127054217 | 0.33 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr19_-_40324255 | 0.33 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr2_+_155554797 | 0.33 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr19_+_50270219 | 0.32 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr2_+_134877740 | 0.32 |
ENST00000409645.1
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr15_-_86338100 | 0.32 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr1_+_200860122 | 0.32 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr17_-_34890759 | 0.32 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr19_+_2476116 | 0.32 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr16_-_31076332 | 0.32 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr2_-_31361362 | 0.31 |
ENST00000430167.1
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr4_-_7873981 | 0.31 |
ENST00000360265.4
|
AFAP1
|
actin filament associated protein 1 |
| chr10_+_11784360 | 0.31 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr11_+_65480222 | 0.31 |
ENST00000534681.1
|
KAT5
|
K(lysine) acetyltransferase 5 |
| chr11_-_65381643 | 0.31 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr8_+_21912328 | 0.31 |
ENST00000432128.1
ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN
|
dematin actin binding protein |
| chr12_+_7033616 | 0.31 |
ENST00000356654.4
|
ATN1
|
atrophin 1 |
| chr20_+_62666902 | 0.30 |
ENST00000431158.1
|
LINC00176
|
long intergenic non-protein coding RNA 176 |
| chr1_-_11751665 | 0.30 |
ENST00000376667.3
ENST00000235310.3 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chrX_+_100878112 | 0.30 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr2_+_27070964 | 0.30 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr2_+_128177458 | 0.30 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr1_-_206785789 | 0.30 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr19_+_47105309 | 0.30 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr16_+_31085714 | 0.30 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr13_-_74993252 | 0.30 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr6_+_32936942 | 0.29 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr2_+_219246746 | 0.29 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr5_+_176560742 | 0.29 |
ENST00000439151.2
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr1_-_11751529 | 0.29 |
ENST00000376672.1
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr16_+_77225071 | 0.29 |
ENST00000439557.2
ENST00000545553.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr2_-_27632390 | 0.29 |
ENST00000350803.4
ENST00000344034.4 |
PPM1G
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr6_+_43737939 | 0.29 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chrX_-_46759138 | 0.29 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31 |
| chr11_-_72432950 | 0.29 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_-_120805872 | 0.29 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr5_-_172198190 | 0.29 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr3_-_52273098 | 0.29 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr1_-_247495045 | 0.29 |
ENST00000294753.4
ENST00000366498.2 |
ZNF496
|
zinc finger protein 496 |
| chr11_-_129872672 | 0.29 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr17_-_73874654 | 0.29 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr12_-_118490403 | 0.29 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr16_-_30381580 | 0.29 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr10_+_133918175 | 0.28 |
ENST00000298622.4
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr3_+_32280159 | 0.28 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr11_+_75273246 | 0.28 |
ENST00000526397.1
ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr14_+_103394963 | 0.28 |
ENST00000559525.1
ENST00000559789.1 |
AMN
|
amnion associated transmembrane protein |
| chr1_-_1310530 | 0.28 |
ENST00000338370.3
ENST00000321751.5 ENST00000378853.3 |
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr16_+_72088376 | 0.28 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr16_+_72142195 | 0.28 |
ENST00000563819.1
ENST00000567142.2 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr17_-_40075197 | 0.28 |
ENST00000590770.1
ENST00000590151.1 |
ACLY
|
ATP citrate lyase |
| chr1_-_154943002 | 0.28 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr7_-_73133959 | 0.28 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr19_-_5624057 | 0.28 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr15_+_85923856 | 0.28 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr7_-_150946015 | 0.27 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr7_-_150974494 | 0.27 |
ENST00000392811.2
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr2_-_98950434 | 0.27 |
ENST00000409490.1
|
AC092675.3
|
Uncharacterized protein |
| chr17_+_38465441 | 0.27 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr5_-_54523143 | 0.27 |
ENST00000513312.1
|
MCIDAS
|
multiciliate differentiation and DNA synthesis associated cell cycle protein |
| chr10_+_114710516 | 0.27 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_113247543 | 0.27 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr3_-_49170522 | 0.27 |
ENST00000418109.1
|
LAMB2
|
laminin, beta 2 (laminin S) |
| chr17_-_34890732 | 0.27 |
ENST00000268852.9
|
MYO19
|
myosin XIX |
| chr16_+_57126482 | 0.27 |
ENST00000537605.1
ENST00000535318.2 |
CPNE2
|
copine II |
| chr19_+_2164126 | 0.27 |
ENST00000398665.3
|
DOT1L
|
DOT1-like histone H3K79 methyltransferase |
| chr7_+_75932863 | 0.26 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr1_+_206623784 | 0.26 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr19_-_56092187 | 0.26 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chr11_+_75273101 | 0.26 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr11_-_8739383 | 0.26 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr16_+_67233412 | 0.26 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chr22_-_19466454 | 0.26 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr8_+_23145594 | 0.26 |
ENST00000519952.1
ENST00000518840.1 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.2 | 0.7 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 0.7 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.2 | 0.9 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 1.9 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.2 | 0.5 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.4 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.5 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.1 | 0.8 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.1 | 0.4 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.7 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.4 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.3 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.2 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.5 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.7 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.4 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) L-arginine import(GO:0043091) divalent metal ion export(GO:0070839) arginine import(GO:0090467) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.3 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.1 | 0.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.3 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.3 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.5 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.1 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.5 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.7 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 0.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.2 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.2 | GO:0021557 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.6 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.2 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.6 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.6 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 1.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 1.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.4 | GO:2000553 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.9 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.3 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.7 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.6 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.1 | GO:0061088 | cobalt ion transport(GO:0006824) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 1.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 0.7 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 0.5 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.8 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.2 | 0.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.5 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.2 | 0.5 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.4 | GO:0016826 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.1 | 0.4 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.6 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.6 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.2 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.3 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.5 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 1.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |