Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
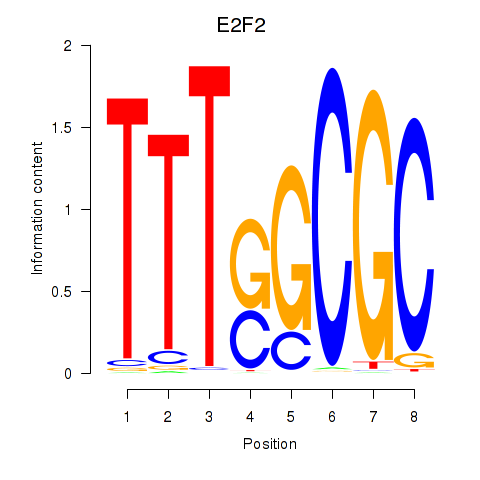
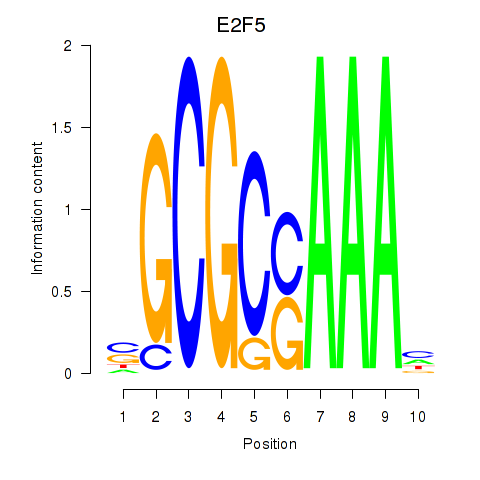
Results for E2F2_E2F5
Z-value: 1.09
Transcription factors associated with E2F2_E2F5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F2
|
ENSG00000007968.6 | E2F transcription factor 2 |
|
E2F5
|
ENSG00000133740.6 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F5 | hg19_v2_chr8_+_86099884_86099920 | 0.88 | 2.0e-02 | Click! |
| E2F2 | hg19_v2_chr1_-_23857698_23857733 | -0.83 | 3.9e-02 | Click! |
Activity profile of E2F2_E2F5 motif
Sorted Z-values of E2F2_E2F5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_17935119 | 1.36 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr10_+_62538248 | 1.23 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr2_-_17935059 | 1.09 |
ENST00000448223.2
ENST00000381272.4 ENST00000351948.4 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr7_+_26241310 | 1.08 |
ENST00000396386.2
|
CBX3
|
chromobox homolog 3 |
| chr11_-_95522639 | 1.06 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr19_+_36705504 | 1.06 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr19_+_36706024 | 1.02 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chr2_-_17935027 | 1.02 |
ENST00000446852.1
|
SMC6
|
structural maintenance of chromosomes 6 |
| chr10_+_62538089 | 0.88 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr7_+_26241325 | 0.88 |
ENST00000456948.1
ENST00000409747.1 |
CBX3
|
chromobox homolog 3 |
| chr10_+_112327425 | 0.87 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr11_-_95522907 | 0.83 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr14_+_36295638 | 0.81 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr14_-_50154921 | 0.73 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr20_-_35724388 | 0.73 |
ENST00000344359.3
ENST00000373664.3 |
RBL1
|
retinoblastoma-like 1 (p107) |
| chr8_-_124408652 | 0.70 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr17_+_29158962 | 0.69 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr6_+_24775641 | 0.67 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr15_+_71185148 | 0.67 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_-_203103281 | 0.65 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr1_-_26232522 | 0.65 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr5_+_36876833 | 0.64 |
ENST00000282516.8
ENST00000448238.2 |
NIPBL
|
Nipped-B homolog (Drosophila) |
| chr6_+_27114861 | 0.63 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chrX_+_123095546 | 0.59 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr1_-_91487770 | 0.59 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr14_+_36295504 | 0.58 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr22_-_38966172 | 0.56 |
ENST00000216024.2
|
DMC1
|
DNA meiotic recombinase 1 |
| chr3_+_44803322 | 0.56 |
ENST00000481166.2
|
KIF15
|
kinesin family member 15 |
| chr6_+_24775153 | 0.55 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr15_-_66649010 | 0.54 |
ENST00000367709.4
ENST00000261881.4 |
TIPIN
|
TIMELESS interacting protein |
| chr10_+_96305610 | 0.54 |
ENST00000371332.4
ENST00000239026.6 |
HELLS
|
helicase, lymphoid-specific |
| chr15_+_44719394 | 0.53 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr7_+_86781677 | 0.53 |
ENST00000331242.7
ENST00000394702.3 ENST00000413276.2 ENST00000446796.2 ENST00000411766.2 ENST00000420131.1 ENST00000414630.2 ENST00000453049.1 ENST00000428819.1 ENST00000448598.1 ENST00000449088.3 ENST00000430405.3 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr8_-_120868078 | 0.53 |
ENST00000313655.4
|
DSCC1
|
DNA replication and sister chromatid cohesion 1 |
| chr2_+_48010312 | 0.52 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr15_-_71184724 | 0.52 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr15_+_71184931 | 0.51 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_-_47168850 | 0.51 |
ENST00000409207.1
|
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr21_-_30365136 | 0.51 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr13_+_32889605 | 0.51 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chr1_-_26232951 | 0.50 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr1_-_91487013 | 0.49 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr1_-_100598444 | 0.49 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr4_+_71859156 | 0.49 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr10_-_77161004 | 0.49 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chr1_-_222885770 | 0.48 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr5_+_79950463 | 0.46 |
ENST00000265081.6
|
MSH3
|
mutS homolog 3 |
| chr7_-_94285472 | 0.45 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr6_+_27782788 | 0.45 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr2_+_48010221 | 0.44 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6 |
| chr20_+_5931275 | 0.44 |
ENST00000378896.3
ENST00000378883.1 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr5_+_56469843 | 0.43 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr4_+_154265784 | 0.43 |
ENST00000240488.3
|
MND1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr10_+_96305535 | 0.43 |
ENST00000419900.1
ENST00000348459.5 ENST00000394045.1 ENST00000394044.1 ENST00000394036.1 |
HELLS
|
helicase, lymphoid-specific |
| chrX_+_24711997 | 0.43 |
ENST00000379068.3
ENST00000379059.3 |
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr5_-_36152031 | 0.42 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr7_+_26240776 | 0.41 |
ENST00000337620.4
|
CBX3
|
chromobox homolog 3 |
| chr6_+_27775899 | 0.41 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr2_-_47168906 | 0.41 |
ENST00000444761.2
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr7_-_94285402 | 0.40 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chrX_+_123095890 | 0.40 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr22_+_42394780 | 0.40 |
ENST00000328823.9
|
WBP2NL
|
WBP2 N-terminal like |
| chr15_+_44719970 | 0.38 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr8_+_94929077 | 0.38 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_+_222885884 | 0.37 |
ENST00000340934.5
|
BROX
|
BRO1 domain and CAAX motif containing |
| chr7_+_86781847 | 0.37 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr13_+_45694583 | 0.36 |
ENST00000340473.6
|
GTF2F2
|
general transcription factor IIF, polypeptide 2, 30kDa |
| chr17_+_30264014 | 0.36 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr13_-_73356009 | 0.36 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr5_+_93954358 | 0.35 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr1_-_26233423 | 0.35 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chrX_+_123095860 | 0.34 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr3_+_44803209 | 0.34 |
ENST00000326047.4
|
KIF15
|
kinesin family member 15 |
| chr8_+_94929168 | 0.33 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr2_-_55277692 | 0.33 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr20_+_5931497 | 0.33 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr5_+_56469939 | 0.32 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr15_-_64673665 | 0.32 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr2_+_17935383 | 0.32 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr20_-_5931109 | 0.32 |
ENST00000203001.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr3_-_52719888 | 0.31 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr5_-_79950371 | 0.31 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chr9_+_17134980 | 0.30 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr15_-_64673630 | 0.30 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr5_+_93954039 | 0.29 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr1_+_6845384 | 0.29 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr6_+_27833034 | 0.29 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr1_+_25598872 | 0.29 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr2_+_170440844 | 0.29 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr11_+_9406169 | 0.29 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr6_+_88299833 | 0.28 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr1_-_205719295 | 0.28 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr2_-_219433014 | 0.27 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr17_+_42733803 | 0.27 |
ENST00000409122.2
|
C17orf104
|
chromosome 17 open reading frame 104 |
| chr7_-_27169801 | 0.27 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr9_+_17135016 | 0.27 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr15_+_32907691 | 0.27 |
ENST00000361627.3
ENST00000567348.1 ENST00000563864.1 ENST00000543522.1 |
ARHGAP11A
|
Rho GTPase activating protein 11A |
| chr5_+_56469775 | 0.27 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr5_-_74062867 | 0.27 |
ENST00000509097.1
|
GFM2
|
G elongation factor, mitochondrial 2 |
| chr19_-_409134 | 0.26 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chr7_+_86781778 | 0.26 |
ENST00000432937.2
|
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr7_-_94285511 | 0.26 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr16_-_30102547 | 0.26 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr9_+_106856541 | 0.26 |
ENST00000286398.7
ENST00000440179.1 ENST00000374793.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chrX_-_122866874 | 0.26 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr9_+_106856831 | 0.25 |
ENST00000303219.8
ENST00000374787.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr3_+_182511266 | 0.25 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr11_+_125495862 | 0.25 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
| chr8_+_94929273 | 0.25 |
ENST00000518573.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr2_+_170440902 | 0.24 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr4_-_170679024 | 0.24 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr15_+_44719790 | 0.24 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr18_+_2655849 | 0.24 |
ENST00000261598.8
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr4_+_178230985 | 0.23 |
ENST00000264596.3
|
NEIL3
|
nei endonuclease VIII-like 3 (E. coli) |
| chr17_-_17184546 | 0.23 |
ENST00000417352.1
|
COPS3
|
COP9 signalosome subunit 3 |
| chr22_+_35653445 | 0.23 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr9_+_86595626 | 0.23 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr11_-_95523500 | 0.22 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr3_-_10028366 | 0.22 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr2_-_203103185 | 0.22 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chrX_+_131157322 | 0.22 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr1_-_23670817 | 0.22 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_6845497 | 0.22 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr1_-_47779762 | 0.22 |
ENST00000371877.3
ENST00000360380.3 ENST00000337817.5 ENST00000447475.2 |
STIL
|
SCL/TAL1 interrupting locus |
| chr15_+_100106155 | 0.22 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr10_-_25241499 | 0.21 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr1_+_13910194 | 0.21 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr4_-_18023350 | 0.21 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr15_+_44719996 | 0.21 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_+_212208919 | 0.21 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr4_+_129730947 | 0.21 |
ENST00000452328.2
ENST00000504089.1 |
PHF17
|
jade family PHD finger 1 |
| chr6_-_88299678 | 0.20 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr7_-_105516923 | 0.20 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr12_-_31479045 | 0.20 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr18_+_19192228 | 0.20 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr5_-_137548997 | 0.20 |
ENST00000505120.1
ENST00000394886.2 ENST00000394884.3 |
CDC23
|
cell division cycle 23 |
| chr22_-_29137771 | 0.20 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr3_+_41240925 | 0.20 |
ENST00000396183.3
ENST00000349496.5 ENST00000453024.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr1_+_66458072 | 0.19 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chrX_+_131157290 | 0.19 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr17_+_42733730 | 0.19 |
ENST00000359945.3
ENST00000425535.1 |
C17orf104
|
chromosome 17 open reading frame 104 |
| chr11_+_74303575 | 0.19 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr14_-_69864993 | 0.19 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr7_+_6048856 | 0.19 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr3_+_14219858 | 0.19 |
ENST00000306024.3
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_23670813 | 0.19 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr7_-_158497431 | 0.19 |
ENST00000449727.2
ENST00000409339.3 ENST00000409423.1 ENST00000356309.3 |
NCAPG2
|
non-SMC condensin II complex, subunit G2 |
| chr7_+_94139105 | 0.18 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr18_-_54318353 | 0.18 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr1_-_23670752 | 0.18 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr4_+_129730779 | 0.18 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr17_+_57642886 | 0.18 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr6_-_26043885 | 0.18 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chrX_-_106243451 | 0.18 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr8_-_95565673 | 0.18 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr2_+_232573208 | 0.18 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr11_-_19262486 | 0.17 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chr11_+_9595180 | 0.17 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr1_+_27113963 | 0.17 |
ENST00000430292.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr2_-_54014055 | 0.17 |
ENST00000263634.3
ENST00000406687.1 |
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chr6_+_4021554 | 0.17 |
ENST00000337659.6
|
PRPF4B
|
pre-mRNA processing factor 4B |
| chr18_-_70210764 | 0.16 |
ENST00000585159.1
ENST00000584764.1 |
CBLN2
|
cerebellin 2 precursor |
| chr1_+_222988464 | 0.16 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr12_+_27932803 | 0.16 |
ENST00000381271.2
|
KLHL42
|
kelch-like family member 42 |
| chr1_-_54303934 | 0.16 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr3_-_133380731 | 0.16 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr19_-_36705763 | 0.16 |
ENST00000591473.1
|
ZNF565
|
zinc finger protein 565 |
| chr15_+_100106244 | 0.16 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr1_-_91487806 | 0.15 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr19_-_10305302 | 0.15 |
ENST00000592054.1
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr5_+_43120985 | 0.15 |
ENST00000515326.1
|
ZNF131
|
zinc finger protein 131 |
| chr2_-_55277436 | 0.15 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chrX_+_100353153 | 0.15 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr1_-_24306768 | 0.15 |
ENST00000374453.3
ENST00000453840.3 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr1_+_33116743 | 0.15 |
ENST00000414241.3
ENST00000373493.5 |
RBBP4
|
retinoblastoma binding protein 4 |
| chr14_+_35452104 | 0.15 |
ENST00000216774.6
ENST00000546080.1 |
SRP54
|
signal recognition particle 54kDa |
| chr2_+_54014168 | 0.15 |
ENST00000405123.3
ENST00000185150.4 ENST00000378239.5 |
ERLEC1
|
endoplasmic reticulum lectin 1 |
| chr8_-_98290087 | 0.15 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr2_-_54014127 | 0.15 |
ENST00000394717.2
|
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chr6_-_160210604 | 0.14 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr2_+_47630108 | 0.14 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr3_-_52719912 | 0.14 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chrX_+_21958814 | 0.14 |
ENST00000379404.1
ENST00000415881.2 |
SMS
|
spermine synthase |
| chr2_-_100106419 | 0.14 |
ENST00000393445.3
ENST00000258428.3 |
REV1
|
REV1, polymerase (DNA directed) |
| chrX_+_70752917 | 0.13 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr21_+_17961006 | 0.13 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_+_171571827 | 0.13 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr5_+_74062806 | 0.13 |
ENST00000296802.5
|
NSA2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chrX_+_46696372 | 0.13 |
ENST00000218340.3
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr2_+_16080659 | 0.13 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr6_-_160210715 | 0.13 |
ENST00000392168.2
ENST00000321394.7 |
TCP1
|
t-complex 1 |
| chr2_-_235405679 | 0.13 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chrX_-_106243294 | 0.13 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr11_+_4116054 | 0.13 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr2_-_136633940 | 0.13 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr16_-_47495170 | 0.12 |
ENST00000320640.6
ENST00000544001.2 |
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr1_+_100598691 | 0.12 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr3_-_136471204 | 0.12 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr1_+_35734616 | 0.12 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr6_-_17706618 | 0.12 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F2_E2F5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.3 | 2.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.3 | 0.9 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.3 | 0.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 0.3 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.2 | 1.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.6 | GO:0061010 | external genitalia morphogenesis(GO:0035261) gall bladder development(GO:0061010) |
| 0.2 | 1.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 2.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.4 | GO:0007343 | egg activation(GO:0007343) female pronucleus assembly(GO:0035038) |
| 0.1 | 1.6 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.4 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0042000 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.7 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 1.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.5 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.2 | GO:1904793 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.5 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 1.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.5 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.0 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.4 | 2.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.3 | 2.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 0.9 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.2 | 1.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.8 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 1.6 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.2 | 2.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 1.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.9 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.3 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 1.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 2.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.5 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 3.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |