Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
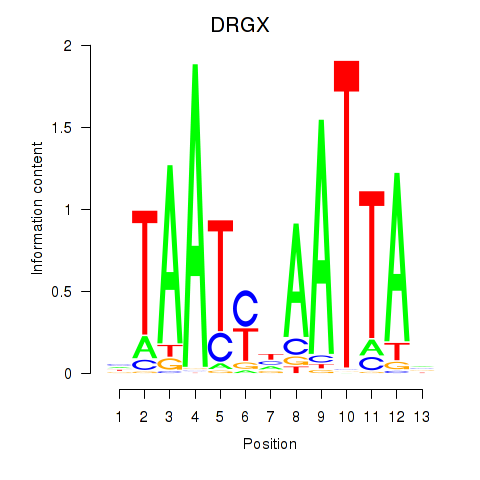
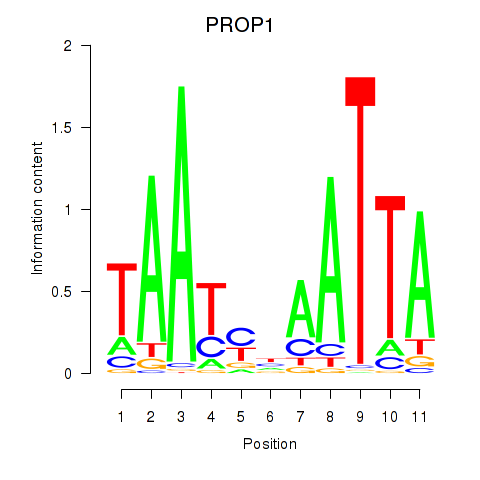
Results for DRGX_PROP1
Z-value: 0.46
Transcription factors associated with DRGX_PROP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DRGX
|
ENSG00000165606.4 | dorsal root ganglia homeobox |
|
PROP1
|
ENSG00000175325.2 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DRGX | hg19_v2_chr10_-_50603497_50603497 | 0.24 | 6.5e-01 | Click! |
Activity profile of DRGX_PROP1 motif
Sorted Z-values of DRGX_PROP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_81801115 | 0.45 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr11_-_111649074 | 0.28 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr15_+_66874502 | 0.28 |
ENST00000558797.1
|
RP11-321F6.1
|
HCG2003567; Uncharacterized protein |
| chr12_+_2912363 | 0.25 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr3_-_186262166 | 0.25 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr1_+_50459990 | 0.23 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr1_-_151148442 | 0.23 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr3_+_111393501 | 0.23 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr18_+_20714525 | 0.22 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr3_-_195310802 | 0.19 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_+_39944025 | 0.19 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr1_-_151148492 | 0.17 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr3_+_138340067 | 0.17 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_+_77318769 | 0.17 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chrX_+_107288239 | 0.17 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr16_+_85932760 | 0.16 |
ENST00000565552.1
|
IRF8
|
interferon regulatory factor 8 |
| chr3_-_191000172 | 0.16 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chrX_+_107288197 | 0.16 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_191334212 | 0.16 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr1_-_94147385 | 0.16 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_29923893 | 0.16 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr9_-_71869642 | 0.15 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr14_-_95236551 | 0.15 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr17_-_61045902 | 0.15 |
ENST00000581596.1
|
RP11-180P8.3
|
RP11-180P8.3 |
| chr12_-_118797475 | 0.15 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr19_-_58485895 | 0.15 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr5_-_111093759 | 0.14 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr12_-_118796971 | 0.14 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chrX_-_15332665 | 0.14 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr7_+_107224364 | 0.13 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_-_234667504 | 0.13 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr10_-_25304889 | 0.13 |
ENST00000483339.2
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr6_+_15401075 | 0.13 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr11_+_1295809 | 0.12 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr13_-_47471155 | 0.12 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chrX_-_19817869 | 0.12 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr15_-_44092295 | 0.12 |
ENST00000249714.3
ENST00000299969.6 ENST00000319327.6 |
SERINC4
|
serine incorporator 4 |
| chrX_-_11284095 | 0.12 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr1_-_232651312 | 0.12 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_-_107235378 | 0.11 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr6_-_32908765 | 0.11 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_+_101293687 | 0.10 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr2_+_102413726 | 0.10 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr14_+_38264418 | 0.10 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr6_-_107235287 | 0.10 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr17_-_14683517 | 0.10 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr3_-_196242233 | 0.10 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_62439037 | 0.10 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr16_+_81272287 | 0.10 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr3_+_138340049 | 0.10 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chrX_+_18725758 | 0.10 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr1_-_21377383 | 0.10 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr22_-_43398442 | 0.09 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_+_27791862 | 0.09 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr3_-_9291063 | 0.09 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr1_+_243419306 | 0.09 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr18_+_61445205 | 0.09 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr5_+_142149932 | 0.09 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr6_-_41039567 | 0.09 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr5_+_59783540 | 0.09 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_+_54422142 | 0.09 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr17_-_39623681 | 0.09 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr5_+_140480083 | 0.08 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr5_+_142149955 | 0.08 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr8_-_95229531 | 0.08 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr3_+_111393659 | 0.08 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr7_+_55980331 | 0.08 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr19_+_13842559 | 0.08 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr2_+_162272605 | 0.08 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr1_+_109756523 | 0.08 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr10_-_61900762 | 0.07 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_15668240 | 0.07 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr14_-_81893734 | 0.07 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr2_+_109237717 | 0.07 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_187455680 | 0.07 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr1_+_206557366 | 0.07 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_+_44588877 | 0.07 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr9_-_39239171 | 0.07 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr18_+_66382428 | 0.07 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr14_-_101295407 | 0.07 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr16_+_68119440 | 0.06 |
ENST00000346183.3
ENST00000329524.4 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr12_+_122688090 | 0.06 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr16_-_21663950 | 0.06 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr12_-_118628350 | 0.06 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr15_+_42066632 | 0.06 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr10_+_695888 | 0.06 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr22_-_29107919 | 0.06 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr6_-_66417107 | 0.06 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chrX_+_100878112 | 0.06 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr16_+_68119247 | 0.06 |
ENST00000575270.1
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr11_-_57479673 | 0.06 |
ENST00000337672.2
ENST00000431606.2 |
MED19
|
mediator complex subunit 19 |
| chr1_-_242162375 | 0.06 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr6_-_32908792 | 0.06 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_+_170632250 | 0.06 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr10_+_24544249 | 0.06 |
ENST00000430453.2
|
KIAA1217
|
KIAA1217 |
| chr5_+_137722255 | 0.06 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr16_+_68119324 | 0.06 |
ENST00000349223.5
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr4_-_76957214 | 0.06 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr9_+_2159850 | 0.06 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_40909354 | 0.06 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr5_-_134735568 | 0.06 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr5_-_136649218 | 0.05 |
ENST00000510405.1
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr1_+_174933899 | 0.05 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_+_55192267 | 0.05 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr9_-_34397800 | 0.05 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr6_-_10435032 | 0.05 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr13_+_88325498 | 0.05 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr10_+_97803151 | 0.05 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr9_+_21440440 | 0.05 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr13_-_95131923 | 0.05 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr1_-_157069590 | 0.05 |
ENST00000454449.2
|
ETV3L
|
ets variant 3-like |
| chr1_+_180199393 | 0.05 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr9_+_42671887 | 0.05 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr5_+_140514782 | 0.05 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr6_+_12007963 | 0.05 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr5_+_59783941 | 0.05 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_+_3773825 | 0.05 |
ENST00000378209.3
ENST00000338895.3 ENST00000378212.2 ENST00000341385.3 |
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
| chr2_+_179184955 | 0.04 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr6_+_25992884 | 0.04 |
ENST00000606723.2
|
U91328.19
|
U91328.19 |
| chrX_+_10126488 | 0.04 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr4_-_184243561 | 0.04 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr6_+_42896865 | 0.04 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr22_-_30642728 | 0.04 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr11_+_108094786 | 0.04 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr12_+_19358228 | 0.04 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr9_+_20927764 | 0.04 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr19_+_47616682 | 0.04 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr3_-_141747950 | 0.04 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr13_-_88463487 | 0.04 |
ENST00000606221.1
|
RP11-471M2.3
|
RP11-471M2.3 |
| chr15_+_51669513 | 0.04 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr6_+_3259122 | 0.04 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr15_+_44719996 | 0.04 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr6_+_26087646 | 0.04 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr6_+_116691001 | 0.04 |
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase |
| chr20_+_32150140 | 0.04 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr4_-_174320687 | 0.04 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr2_+_152214098 | 0.03 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr11_+_8040739 | 0.03 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr6_+_88117683 | 0.03 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr10_-_99030395 | 0.03 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr3_-_11623804 | 0.03 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_-_92371839 | 0.03 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr10_+_89420706 | 0.03 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr4_-_176733897 | 0.03 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr3_+_153839149 | 0.03 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr4_+_142558078 | 0.03 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr7_+_93535817 | 0.03 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr16_+_1728257 | 0.03 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr6_+_3259148 | 0.03 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr11_-_63439013 | 0.03 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr8_+_92114060 | 0.03 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr15_+_75628232 | 0.03 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr13_+_20268547 | 0.03 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr9_-_27005686 | 0.03 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr4_+_88571429 | 0.03 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr4_-_187647773 | 0.03 |
ENST00000509647.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr12_-_110883346 | 0.03 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr3_-_197300194 | 0.02 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr9_+_125795788 | 0.02 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr16_+_69373323 | 0.02 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr5_-_54468974 | 0.02 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr7_+_152456904 | 0.02 |
ENST00000537264.1
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr3_-_156878482 | 0.02 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr3_+_155860751 | 0.02 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr18_-_2982869 | 0.02 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr14_+_21566980 | 0.02 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr16_-_69373396 | 0.02 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr11_-_96076334 | 0.02 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr6_-_24358264 | 0.02 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr21_-_15583165 | 0.02 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr12_+_100897130 | 0.02 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr22_+_25615489 | 0.02 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr7_-_80551671 | 0.02 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr8_+_7801144 | 0.02 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr5_+_140227357 | 0.02 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr14_+_38065052 | 0.02 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr2_+_121010370 | 0.02 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr16_+_81678957 | 0.02 |
ENST00000398040.4
|
CMIP
|
c-Maf inducing protein |
| chr2_-_70529126 | 0.02 |
ENST00000438759.1
ENST00000430566.1 |
FAM136A
|
family with sequence similarity 136, member A |
| chr11_-_111649015 | 0.02 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_-_160279207 | 0.02 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr12_-_15815626 | 0.02 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr3_-_87325728 | 0.02 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr2_+_74685413 | 0.02 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr19_-_11456905 | 0.02 |
ENST00000588560.1
ENST00000592952.1 |
TMEM205
|
transmembrane protein 205 |
| chr2_-_200320768 | 0.02 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chr12_+_54410664 | 0.02 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr17_-_27332931 | 0.02 |
ENST00000442608.3
ENST00000335960.6 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr8_+_67039131 | 0.02 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr9_+_123884038 | 0.02 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr8_-_112248400 | 0.02 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr1_+_84630645 | 0.02 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_57873631 | 0.02 |
ENST00000393791.3
ENST00000356411.2 ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr9_+_12693336 | 0.02 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr6_-_150067696 | 0.02 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr22_+_22723969 | 0.02 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr16_-_67517716 | 0.02 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr11_+_22694123 | 0.01 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr6_+_88106840 | 0.01 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr4_+_156587853 | 0.01 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DRGX_PROP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1903464 | negative regulation of mitotic cell cycle DNA replication(GO:1903464) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |