Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
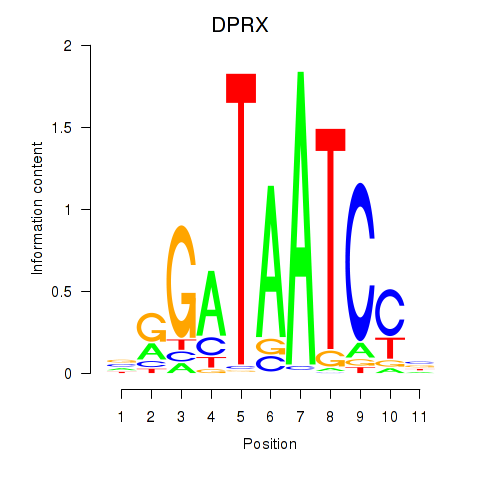
Results for DPRX
Z-value: 1.34
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DPRX | hg19_v2_chr19_+_54135310_54135310 | -0.58 | 2.2e-01 | Click! |
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_321340 | 10.59 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_-_321050 | 5.09 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr12_+_113344755 | 1.19 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_113344582 | 1.12 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_66796401 | 0.73 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr4_+_89299885 | 0.73 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_+_113344811 | 0.67 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_6949964 | 0.64 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr1_-_150208320 | 0.59 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_63461642 | 0.56 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr22_-_38480100 | 0.51 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr5_-_77072145 | 0.45 |
ENST00000380377.4
|
TBCA
|
tubulin folding cofactor A |
| chr20_+_10199566 | 0.45 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr4_-_153700864 | 0.44 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr12_+_104359641 | 0.42 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr18_-_28681950 | 0.41 |
ENST00000251081.6
|
DSC2
|
desmocollin 2 |
| chr12_+_113416265 | 0.40 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr4_-_87374330 | 0.38 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_-_235405168 | 0.37 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr17_-_65992544 | 0.37 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr12_+_104359614 | 0.36 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr8_+_104384616 | 0.36 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr3_-_54962100 | 0.35 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr5_-_54988448 | 0.33 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr1_-_68516393 | 0.33 |
ENST00000395201.1
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3 |
| chr4_-_147443043 | 0.33 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr15_+_42565464 | 0.32 |
ENST00000562170.1
ENST00000562859.1 |
GANC
|
glucosidase, alpha; neutral C |
| chr5_-_77072085 | 0.32 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr1_+_63989004 | 0.31 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr18_-_28682374 | 0.31 |
ENST00000280904.6
|
DSC2
|
desmocollin 2 |
| chr20_-_36794938 | 0.30 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr2_-_119605253 | 0.29 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr8_+_107630340 | 0.29 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr7_+_89975979 | 0.29 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr1_-_209741018 | 0.29 |
ENST00000424696.2
|
RP1-272L16.1
|
RP1-272L16.1 |
| chr6_+_7107999 | 0.28 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr16_+_30386098 | 0.27 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr12_-_104359475 | 0.27 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr17_-_26127525 | 0.27 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chrX_+_53449887 | 0.27 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr15_-_42565221 | 0.26 |
ENST00000563371.1
ENST00000568400.1 ENST00000568432.1 |
TMEM87A
|
transmembrane protein 87A |
| chr3_+_111630451 | 0.26 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr6_+_130339710 | 0.26 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr10_+_70661014 | 0.26 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr17_+_48351785 | 0.25 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr10_+_104178946 | 0.25 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr8_+_35649365 | 0.25 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr5_-_59064458 | 0.25 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_110937351 | 0.25 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr5_+_35856951 | 0.25 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr5_+_138678131 | 0.25 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_+_207262881 | 0.25 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr18_-_32957260 | 0.24 |
ENST00000587422.1
ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396
|
zinc finger protein 396 |
| chr1_-_149400542 | 0.24 |
ENST00000392948.2
|
HIST2H3PS2
|
histone cluster 2, H3, pseudogene 2 |
| chr7_-_37488547 | 0.24 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr20_-_36794902 | 0.24 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr5_-_180235755 | 0.24 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_+_104359576 | 0.24 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr11_-_85430088 | 0.23 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_10068095 | 0.23 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr2_+_103378472 | 0.22 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr7_+_73868220 | 0.22 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr3_+_37035289 | 0.22 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr5_+_94955782 | 0.21 |
ENST00000380007.2
|
GPR150
|
G protein-coupled receptor 150 |
| chr6_-_84418432 | 0.21 |
ENST00000519825.1
ENST00000523484.2 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr1_-_150780757 | 0.21 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr11_-_85430204 | 0.21 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr2_+_232063436 | 0.21 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr6_-_37225391 | 0.21 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr6_-_37225367 | 0.21 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr2_+_7118755 | 0.20 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr20_-_34330129 | 0.20 |
ENST00000397370.3
ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39
|
RNA binding motif protein 39 |
| chr1_+_15986364 | 0.20 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr7_-_75401513 | 0.19 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr20_-_43936937 | 0.19 |
ENST00000342716.4
ENST00000353917.5 ENST00000360607.6 ENST00000372751.4 |
MATN4
|
matrilin 4 |
| chr11_-_8816375 | 0.19 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr2_+_27799389 | 0.19 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr4_+_84377198 | 0.19 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr2_+_120517174 | 0.19 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr5_+_118604439 | 0.18 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr9_-_123812542 | 0.18 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr14_+_59951161 | 0.18 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr12_-_56105880 | 0.18 |
ENST00000557257.1
|
ITGA7
|
integrin, alpha 7 |
| chrX_-_15683147 | 0.18 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr1_+_81771806 | 0.18 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr3_-_158390282 | 0.18 |
ENST00000264265.3
|
LXN
|
latexin |
| chr5_+_132083106 | 0.18 |
ENST00000378731.1
|
CCNI2
|
cyclin I family, member 2 |
| chr8_-_65730127 | 0.17 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr7_+_94536898 | 0.17 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr5_-_124081008 | 0.17 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr3_-_15469045 | 0.17 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr17_-_5323480 | 0.17 |
ENST00000573584.1
|
NUP88
|
nucleoporin 88kDa |
| chr7_-_30066233 | 0.17 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr15_-_42565606 | 0.17 |
ENST00000307216.6
ENST00000448392.1 |
TMEM87A
|
transmembrane protein 87A |
| chr5_+_138677515 | 0.17 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr5_+_118604385 | 0.17 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr15_+_63481668 | 0.16 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr4_+_170541835 | 0.16 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr21_-_38362497 | 0.16 |
ENST00000427746.1
ENST00000336648.4 |
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr4_+_159131596 | 0.16 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr11_-_118436606 | 0.16 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_+_35220613 | 0.16 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr19_+_36157715 | 0.16 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr1_-_152086556 | 0.16 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr3_+_130650738 | 0.16 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_+_7108210 | 0.16 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_207262170 | 0.15 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr14_-_50319482 | 0.15 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr10_+_74451883 | 0.15 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr2_+_120517717 | 0.14 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr11_-_61849656 | 0.14 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr20_+_22034809 | 0.14 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr14_-_50319758 | 0.14 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr12_+_133033703 | 0.14 |
ENST00000542627.1
|
RP11-503G7.1
|
RP11-503G7.1 |
| chr4_-_147442817 | 0.14 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr12_+_130554803 | 0.14 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
| chr10_-_103578182 | 0.14 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr3_+_23847432 | 0.14 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr11_-_111944704 | 0.14 |
ENST00000532211.1
|
PIH1D2
|
PIH1 domain containing 2 |
| chr3_+_141105705 | 0.14 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr11_-_85430163 | 0.13 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_159131630 | 0.13 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chrX_-_133931164 | 0.13 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr9_+_35792151 | 0.13 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr3_+_101659682 | 0.13 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr14_+_75469606 | 0.13 |
ENST00000266126.5
|
EIF2B2
|
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr1_+_36772691 | 0.12 |
ENST00000312808.4
ENST00000505871.1 |
SH3D21
|
SH3 domain containing 21 |
| chr11_-_118436707 | 0.12 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr3_+_148447887 | 0.12 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chrX_+_53449839 | 0.12 |
ENST00000457095.1
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr8_-_53626974 | 0.12 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr7_+_73868439 | 0.12 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr3_+_133502877 | 0.12 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr1_+_179561011 | 0.12 |
ENST00000294848.8
ENST00000444136.1 |
TDRD5
|
tudor domain containing 5 |
| chr7_+_135242652 | 0.12 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr5_-_159846066 | 0.12 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr1_+_162351503 | 0.12 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr19_+_50528971 | 0.12 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr1_-_36615065 | 0.12 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr2_+_114737146 | 0.12 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chr11_-_104905840 | 0.12 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr6_+_43215727 | 0.12 |
ENST00000304139.5
|
TTBK1
|
tau tubulin kinase 1 |
| chr11_+_107879459 | 0.12 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr4_-_103998439 | 0.12 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr17_-_7080227 | 0.11 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr3_+_37035263 | 0.11 |
ENST00000458205.2
ENST00000539477.1 |
MLH1
|
mutL homolog 1 |
| chr10_-_89577910 | 0.11 |
ENST00000308448.7
ENST00000541004.1 |
ATAD1
|
ATPase family, AAA domain containing 1 |
| chr12_-_95010147 | 0.11 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr1_-_184943610 | 0.11 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr17_+_7482785 | 0.11 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr7_-_99573677 | 0.11 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr20_+_33759854 | 0.11 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr3_+_52448539 | 0.11 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr14_+_70918874 | 0.11 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr1_-_115301235 | 0.11 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr6_+_31554826 | 0.11 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr6_-_32784687 | 0.11 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr4_-_87279641 | 0.11 |
ENST00000512689.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr9_+_17135016 | 0.11 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chrX_-_77150911 | 0.10 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr6_-_32908765 | 0.10 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr8_-_117886955 | 0.10 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr1_+_29063119 | 0.10 |
ENST00000474884.1
ENST00000542507.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr7_+_134576317 | 0.10 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr1_+_182808474 | 0.10 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr3_+_188664988 | 0.10 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr17_-_37557846 | 0.10 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr4_-_122791583 | 0.10 |
ENST00000506636.1
ENST00000264499.4 |
BBS7
|
Bardet-Biedl syndrome 7 |
| chr3_+_49058444 | 0.10 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr3_+_97483366 | 0.10 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr11_-_111944895 | 0.10 |
ENST00000431456.1
ENST00000280350.4 ENST00000530641.1 |
PIH1D2
|
PIH1 domain containing 2 |
| chr4_-_170924888 | 0.10 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr15_+_42565393 | 0.10 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr1_-_153521714 | 0.10 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr4_-_16077741 | 0.09 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr14_-_22005062 | 0.09 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_+_58138664 | 0.09 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr6_-_84418738 | 0.09 |
ENST00000519779.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_-_103998060 | 0.09 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr14_-_94789663 | 0.09 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr1_-_151965048 | 0.09 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr11_+_63448918 | 0.09 |
ENST00000341307.2
ENST00000356000.3 ENST00000542238.1 |
RTN3
|
reticulon 3 |
| chr6_-_35480640 | 0.09 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr2_-_85555355 | 0.08 |
ENST00000282120.2
ENST00000398263.2 |
TGOLN2
|
trans-golgi network protein 2 |
| chr2_-_68290106 | 0.08 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr6_-_35480705 | 0.08 |
ENST00000229771.6
|
TULP1
|
tubby like protein 1 |
| chr6_+_31555045 | 0.08 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr9_+_17134980 | 0.08 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr11_+_2323349 | 0.08 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chr10_-_103578162 | 0.08 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr11_+_108094786 | 0.08 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr1_-_65533390 | 0.08 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr2_-_85555385 | 0.08 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr6_+_42018614 | 0.08 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr7_+_134464376 | 0.08 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr2_+_26915584 | 0.08 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr11_-_85430356 | 0.08 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr20_-_3140490 | 0.07 |
ENST00000449731.1
ENST00000380266.3 |
UBOX5
FASTKD5
|
U-box domain containing 5 FAST kinase domains 5 |
| chr18_+_20494078 | 0.07 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr8_+_110656344 | 0.07 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 1.0 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.3 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.5 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.5 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 1.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.4 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.3 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 2.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:1903181 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0035696 | egg activation(GO:0007343) monocyte extravasation(GO:0035696) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:2000523 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 16.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.0 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 3.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.3 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.4 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) triplex DNA binding(GO:0045142) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 19.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |