Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
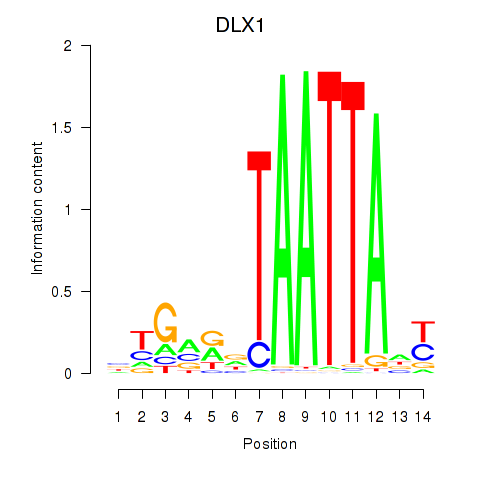
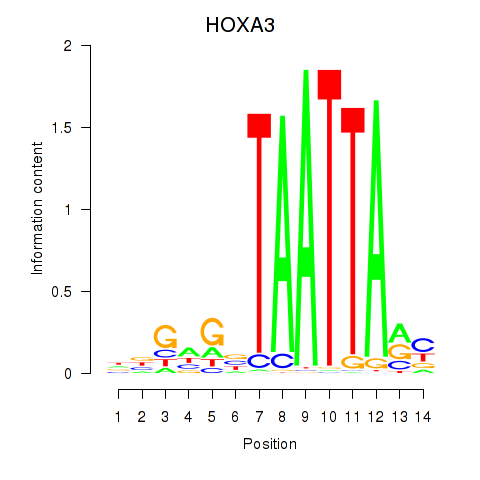
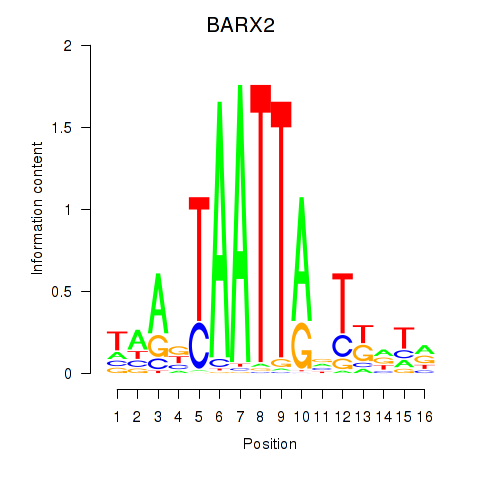
Results for DLX1_HOXA3_BARX2
Z-value: 0.68
Transcription factors associated with DLX1_HOXA3_BARX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX1
|
ENSG00000144355.10 | distal-less homeobox 1 |
|
HOXA3
|
ENSG00000105997.18 | homeobox A3 |
|
BARX2
|
ENSG00000043039.5 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX1 | hg19_v2_chr2_+_172949468_172949522 | 0.80 | 5.8e-02 | Click! |
| HOXA3 | hg19_v2_chr7_-_27153454_27153469 | 0.38 | 4.5e-01 | Click! |
| BARX2 | hg19_v2_chr11_+_129245796_129245835 | -0.12 | 8.2e-01 | Click! |
Activity profile of DLX1_HOXA3_BARX2 motif
Sorted Z-values of DLX1_HOXA3_BARX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_53412368 | 0.44 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr4_-_120243545 | 0.41 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr11_-_63376013 | 0.39 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr12_-_10978957 | 0.30 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr11_-_111649074 | 0.29 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr14_-_30766223 | 0.29 |
ENST00000549360.1
ENST00000508469.2 |
CTD-2251F13.1
|
CTD-2251F13.1 |
| chrX_+_150565653 | 0.28 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr11_+_101918153 | 0.28 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr15_+_58702742 | 0.28 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr4_+_159727272 | 0.27 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr2_-_112237835 | 0.27 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr1_+_225600404 | 0.26 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr11_-_60010556 | 0.26 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr6_-_109702885 | 0.25 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr13_+_111748183 | 0.25 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr17_+_74846230 | 0.25 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr1_-_197115818 | 0.25 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr16_-_25122735 | 0.24 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr14_-_45252031 | 0.23 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr4_+_169013666 | 0.23 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_+_28390637 | 0.22 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr1_+_212965170 | 0.22 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr5_+_136070614 | 0.21 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr19_+_21324827 | 0.20 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr2_-_203735484 | 0.20 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr3_-_98235962 | 0.20 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr11_+_115498761 | 0.20 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr19_+_54466179 | 0.18 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr15_-_37393406 | 0.18 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr16_+_53133070 | 0.18 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_118398178 | 0.18 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr6_-_116833500 | 0.17 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr4_-_36245561 | 0.17 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_+_21579958 | 0.17 |
ENST00000339914.6
ENST00000599461.1 |
ZNF493
|
zinc finger protein 493 |
| chr6_-_138833630 | 0.16 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr13_+_78315295 | 0.16 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_+_87769459 | 0.15 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr12_-_15374343 | 0.15 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr1_+_207226574 | 0.15 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr6_-_41701898 | 0.15 |
ENST00000433032.1
|
TFEB
|
transcription factor EB |
| chr11_-_111649015 | 0.15 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr17_-_18430160 | 0.15 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr2_+_7073174 | 0.14 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr19_+_9296279 | 0.14 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr14_+_55494323 | 0.14 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr1_+_236958554 | 0.14 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr18_+_61616510 | 0.14 |
ENST00000408945.3
|
HMSD
|
histocompatibility (minor) serpin domain containing |
| chr3_-_112127981 | 0.13 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr15_+_35270552 | 0.13 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr16_+_15489603 | 0.13 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr2_-_44550441 | 0.13 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr4_-_103749179 | 0.13 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_101349823 | 0.13 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr3_-_191000172 | 0.13 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr15_+_62853562 | 0.13 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr1_+_196743943 | 0.13 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr5_+_39105358 | 0.13 |
ENST00000593965.1
|
AC008964.1
|
AC008964.1 |
| chr5_+_140557371 | 0.12 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr1_-_159832438 | 0.12 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr16_-_28937027 | 0.12 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr4_-_103749105 | 0.12 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_69111401 | 0.12 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr16_-_1464688 | 0.12 |
ENST00000389221.4
ENST00000508903.2 ENST00000397462.1 ENST00000301712.5 |
UNKL
|
unkempt family zinc finger-like |
| chr12_+_20963632 | 0.11 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr13_+_78315348 | 0.11 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_-_103749313 | 0.11 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr13_+_110958124 | 0.11 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr3_-_105588231 | 0.11 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr6_+_26365443 | 0.11 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr2_-_203735586 | 0.11 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr5_+_66300464 | 0.11 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_202317815 | 0.11 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_+_153552455 | 0.11 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr14_+_55493920 | 0.11 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr12_-_25348007 | 0.11 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr12_+_20963647 | 0.11 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr15_+_89631381 | 0.11 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr14_+_67291158 | 0.11 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr3_+_195447738 | 0.10 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr4_-_119759795 | 0.10 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr2_-_190446738 | 0.10 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr19_+_9203855 | 0.10 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr17_-_19364269 | 0.10 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr2_+_103089756 | 0.10 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr12_-_86650077 | 0.10 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr17_-_4938712 | 0.10 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr12_-_15114603 | 0.10 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr3_-_105587879 | 0.10 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr11_+_59705928 | 0.10 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr14_-_78083112 | 0.10 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr8_+_98900132 | 0.10 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr14_+_100485712 | 0.09 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr13_+_36050881 | 0.09 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr9_-_5830768 | 0.09 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr2_+_102953608 | 0.09 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr7_+_141478242 | 0.09 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr20_-_50418947 | 0.09 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr4_-_109541610 | 0.09 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr16_+_12059091 | 0.09 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr6_+_26365387 | 0.09 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr11_+_33061543 | 0.09 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr19_-_51920952 | 0.09 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_+_100867694 | 0.09 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr8_-_112248400 | 0.09 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr16_+_15489629 | 0.09 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr14_-_39639523 | 0.08 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr10_+_695888 | 0.08 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr15_+_89631647 | 0.08 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr20_-_56265680 | 0.08 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr15_-_49912987 | 0.08 |
ENST00000560246.1
ENST00000558594.1 |
FAM227B
|
family with sequence similarity 227, member B |
| chr16_-_25122785 | 0.08 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr7_-_35013217 | 0.08 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr7_-_124569864 | 0.08 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr3_+_158519654 | 0.08 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr19_-_36001113 | 0.08 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr19_+_49199209 | 0.08 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr6_+_26440700 | 0.08 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr22_-_32766972 | 0.08 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr20_+_816695 | 0.08 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr5_+_67535647 | 0.08 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr20_-_50418972 | 0.08 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_+_196743912 | 0.08 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr2_-_86850949 | 0.08 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr16_-_28634874 | 0.08 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_-_41356347 | 0.08 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chrX_+_107334895 | 0.08 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr19_-_54824344 | 0.07 |
ENST00000346508.3
ENST00000446712.3 ENST00000432233.3 ENST00000301219.3 |
LILRA5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr4_-_46911223 | 0.07 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr12_+_26164645 | 0.07 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_-_101925055 | 0.07 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr8_-_124553437 | 0.07 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr4_-_185275104 | 0.07 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chrX_-_14047996 | 0.07 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr5_-_139726181 | 0.07 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr13_-_30881134 | 0.07 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr1_-_148025848 | 0.07 |
ENST00000310701.10
ENST00000369219.1 ENST00000444640.1 ENST00000431121.1 ENST00000436356.1 ENST00000448574.1 ENST00000458135.1 ENST00000392972.3 ENST00000426874.1 |
NBPF14
|
neuroblastoma breakpoint family, member 14 |
| chr14_+_20187174 | 0.07 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr19_-_3557570 | 0.07 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_-_175041663 | 0.07 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chrX_+_7137475 | 0.07 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr7_-_140482926 | 0.07 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chrX_+_77166172 | 0.07 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr4_-_74486217 | 0.07 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr15_+_75970672 | 0.07 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr6_-_41701428 | 0.07 |
ENST00000424495.1
ENST00000445700.1 |
TFEB
|
transcription factor EB |
| chr18_+_34124507 | 0.07 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr6_-_97731019 | 0.07 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr11_-_63439013 | 0.07 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr20_+_5731083 | 0.06 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr7_+_138915102 | 0.06 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr1_+_149239529 | 0.06 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_81001398 | 0.06 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr10_-_28571015 | 0.06 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr3_+_43732362 | 0.06 |
ENST00000458276.2
|
ABHD5
|
abhydrolase domain containing 5 |
| chr8_-_93978309 | 0.06 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr4_-_103748880 | 0.06 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_160823040 | 0.06 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr2_-_151395525 | 0.06 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr17_-_39623681 | 0.06 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr4_+_123653807 | 0.06 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_-_242612779 | 0.06 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr6_+_160542821 | 0.06 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr2_-_201753859 | 0.06 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr17_+_67498538 | 0.06 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_110427983 | 0.06 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr12_+_100867486 | 0.06 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_-_82957433 | 0.06 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr12_-_91546926 | 0.06 |
ENST00000550758.1
|
DCN
|
decorin |
| chr8_+_54764346 | 0.06 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr11_-_129062093 | 0.06 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr11_-_46638378 | 0.06 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr7_+_130794878 | 0.06 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr11_+_119076745 | 0.06 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr12_-_92821922 | 0.06 |
ENST00000538965.1
ENST00000378487.2 |
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand |
| chr10_-_27529486 | 0.06 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr9_+_108463234 | 0.06 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr12_+_48592134 | 0.06 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr4_+_88896819 | 0.06 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_+_158493642 | 0.06 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr14_+_32798547 | 0.06 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_84609944 | 0.06 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_+_69857515 | 0.06 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr6_+_3259148 | 0.06 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr6_-_38670897 | 0.05 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr15_+_41913690 | 0.05 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr11_+_76092353 | 0.05 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr10_+_115674530 | 0.05 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr16_-_28621298 | 0.05 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr5_-_41794313 | 0.05 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr3_+_186692745 | 0.05 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_-_15563229 | 0.05 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr6_-_9977801 | 0.05 |
ENST00000316020.6
ENST00000491508.1 |
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr1_+_180601139 | 0.05 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chrX_-_13835147 | 0.05 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr12_-_92536433 | 0.05 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr15_+_45544426 | 0.05 |
ENST00000347644.3
ENST00000560438.1 |
SLC28A2
|
solute carrier family 28 (concentrative nucleoside transporter), member 2 |
| chr18_-_53303123 | 0.05 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr6_+_134758827 | 0.05 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr7_+_129015484 | 0.05 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX1_HOXA3_BARX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0038185 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.6 | GO:0046461 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.0 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |