Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
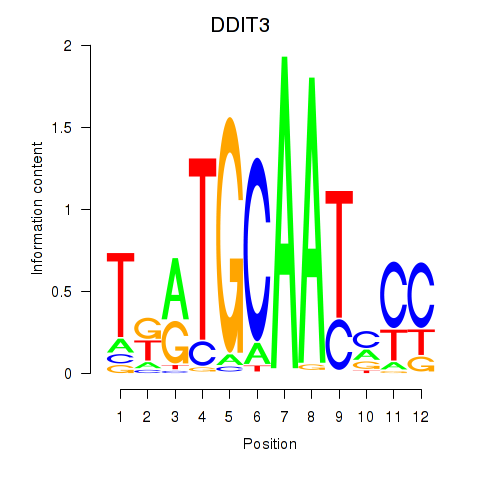
Results for DDIT3
Z-value: 0.63
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.6 | DNA damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DDIT3 | hg19_v2_chr12_-_57914275_57914304 | 0.77 | 7.5e-02 | Click! |
Activity profile of DDIT3 motif
Sorted Z-values of DDIT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_77967433 | 0.79 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr3_-_9595480 | 0.42 |
ENST00000287585.6
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4 |
| chr6_+_144606817 | 0.38 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr2_+_7118755 | 0.36 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr15_+_41245160 | 0.31 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr1_+_149804218 | 0.23 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr14_+_64565442 | 0.22 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr15_+_59730348 | 0.22 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr4_+_157997273 | 0.20 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr12_-_25102252 | 0.20 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chrX_+_16188506 | 0.20 |
ENST00000329538.5
|
MAGEB17
|
melanoma antigen family B, 17 |
| chr1_-_67519782 | 0.19 |
ENST00000235345.5
|
SLC35D1
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr20_+_24929866 | 0.19 |
ENST00000480798.1
ENST00000376835.2 |
CST7
|
cystatin F (leukocystatin) |
| chr15_+_85923797 | 0.18 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr4_+_141178440 | 0.18 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr12_-_25101920 | 0.18 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr14_-_23446003 | 0.18 |
ENST00000553911.1
|
AJUBA
|
ajuba LIM protein |
| chr6_-_48036363 | 0.18 |
ENST00000543600.1
ENST00000398738.2 ENST00000339488.4 |
PTCHD4
|
patched domain containing 4 |
| chr3_-_45883558 | 0.17 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr10_-_17659234 | 0.16 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chrX_-_100129128 | 0.15 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr2_-_159313214 | 0.15 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr7_-_130597935 | 0.14 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr2_-_208030295 | 0.14 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr19_+_1450112 | 0.14 |
ENST00000590469.1
ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2
|
adenomatosis polyposis coli 2 |
| chr2_+_63816126 | 0.13 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr12_-_53074182 | 0.12 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr2_-_43453734 | 0.12 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr2_+_75185619 | 0.12 |
ENST00000483063.1
|
POLE4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr11_+_20044096 | 0.12 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr7_-_56101826 | 0.11 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr9_+_80912059 | 0.11 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr15_+_85923856 | 0.11 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr16_-_18908196 | 0.11 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_114447412 | 0.11 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr19_-_49258606 | 0.11 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr17_-_46691990 | 0.10 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr11_-_8816375 | 0.10 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr12_-_63328817 | 0.10 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr13_-_24007815 | 0.10 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr1_-_161277210 | 0.09 |
ENST00000491222.2
|
MPZ
|
myelin protein zero |
| chr8_+_24151553 | 0.09 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_-_8680383 | 0.09 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr10_-_17659357 | 0.09 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr5_+_112074029 | 0.09 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr1_-_152196669 | 0.08 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr16_-_68269971 | 0.08 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr12_+_64798826 | 0.08 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr10_-_116444371 | 0.08 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr2_-_32390801 | 0.08 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr9_+_70971815 | 0.08 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr16_+_56965960 | 0.08 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr6_-_24489842 | 0.08 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr6_-_157744531 | 0.07 |
ENST00000400788.4
ENST00000367144.4 |
TMEM242
|
transmembrane protein 242 |
| chr11_-_8739566 | 0.07 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr22_+_31518938 | 0.07 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr2_+_63816269 | 0.06 |
ENST00000432309.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr14_-_74226961 | 0.06 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr2_+_63816295 | 0.06 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr17_+_56315936 | 0.06 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr1_+_174844645 | 0.06 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_-_101925055 | 0.06 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr3_+_38017264 | 0.05 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr2_+_223289208 | 0.05 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr4_-_184243561 | 0.05 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr2_+_63816087 | 0.05 |
ENST00000409908.1
ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr15_-_88799948 | 0.05 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr15_-_64126084 | 0.05 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr17_-_34257771 | 0.05 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr14_+_102276132 | 0.04 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr17_+_56315787 | 0.04 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr5_+_72143988 | 0.04 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr9_-_124991124 | 0.04 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr7_+_44040488 | 0.04 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr1_+_90308981 | 0.04 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr1_-_114447683 | 0.04 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr5_+_154135029 | 0.04 |
ENST00000518297.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr17_-_2117600 | 0.04 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr14_+_102276192 | 0.04 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr17_-_77813186 | 0.04 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr2_-_208030647 | 0.03 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_-_104972158 | 0.03 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr8_+_24151620 | 0.03 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr4_-_99578789 | 0.03 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr11_+_62495541 | 0.03 |
ENST00000530625.1
ENST00000513247.2 |
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr19_+_33865218 | 0.03 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr2_-_63815628 | 0.03 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr1_-_207143802 | 0.03 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr1_+_111770294 | 0.03 |
ENST00000474304.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr8_-_18744528 | 0.03 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_+_4336955 | 0.03 |
ENST00000355530.2
|
SPNS3
|
spinster homolog 3 (Drosophila) |
| chr2_-_179672142 | 0.03 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr5_+_145583156 | 0.02 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr10_-_13523073 | 0.02 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr10_-_103874692 | 0.02 |
ENST00000361198.5
|
LDB1
|
LIM domain binding 1 |
| chr11_-_56310757 | 0.02 |
ENST00000528616.2
|
OR5M11
|
olfactory receptor, family 5, subfamily M, member 11 |
| chr20_+_54987168 | 0.02 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr9_+_125486269 | 0.02 |
ENST00000259466.1
|
OR1L4
|
olfactory receptor, family 1, subfamily L, member 4 |
| chr8_-_21669826 | 0.02 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr8_-_11058847 | 0.02 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr1_+_169079823 | 0.02 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr1_+_222910625 | 0.02 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr5_-_131630931 | 0.02 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr7_-_44530479 | 0.02 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr1_+_40840320 | 0.02 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr17_+_7477040 | 0.02 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr14_-_23446021 | 0.02 |
ENST00000553592.1
|
AJUBA
|
ajuba LIM protein |
| chr16_+_56970567 | 0.02 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr14_+_62462541 | 0.01 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr10_+_60028818 | 0.01 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr17_+_4337199 | 0.01 |
ENST00000333476.2
|
SPNS3
|
spinster homolog 3 (Drosophila) |
| chr14_+_32798547 | 0.01 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr7_-_130598059 | 0.01 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr1_+_154229547 | 0.01 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr5_-_66492562 | 0.01 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr3_+_44840679 | 0.01 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr2_-_161350305 | 0.01 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr19_+_41497178 | 0.01 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr2_-_40680578 | 0.00 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_-_31864977 | 0.00 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr18_-_55288973 | 0.00 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chrX_-_138724677 | 0.00 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr8_+_26435915 | 0.00 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr10_+_12391481 | 0.00 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr12_+_50144381 | 0.00 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr22_-_30901637 | 0.00 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr12_+_106994905 | 0.00 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
Network of associatons between targets according to the STRING database.
First level regulatory network of DDIT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.4 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.1 | GO:0010982 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.1 | 0.4 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |