Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
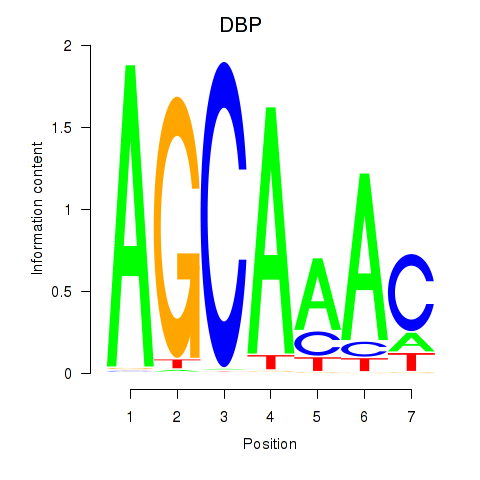
Results for DBP
Z-value: 0.52
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49140692_49140709 | 0.82 | 4.4e-02 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_182850743 | 0.27 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chrX_+_23801280 | 0.26 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr20_+_34020827 | 0.24 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr15_+_78632666 | 0.23 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr2_-_69180083 | 0.22 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr12_+_7169887 | 0.20 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr6_+_26045603 | 0.20 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr7_-_65113280 | 0.19 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr8_-_74495065 | 0.19 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr19_-_56047661 | 0.19 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr14_+_51026844 | 0.19 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr9_+_118916082 | 0.18 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr13_+_44596471 | 0.17 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chr16_+_86612112 | 0.17 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr1_+_114522049 | 0.16 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr1_-_40367668 | 0.16 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr13_+_97928395 | 0.15 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr4_+_95174445 | 0.15 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_155051379 | 0.15 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr18_+_20494078 | 0.15 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr11_+_111411384 | 0.15 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chr1_+_104159999 | 0.14 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr14_+_51026926 | 0.14 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr22_+_45714361 | 0.14 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr20_-_14318248 | 0.14 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr6_-_117747015 | 0.13 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr14_-_92413353 | 0.13 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr8_-_102803163 | 0.13 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr8_+_86999516 | 0.13 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr8_-_40755333 | 0.13 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr16_-_54972581 | 0.13 |
ENST00000559802.1
ENST00000558156.1 |
CTD-3032H12.1
|
CTD-3032H12.1 |
| chr3_-_145878954 | 0.13 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr2_+_210444748 | 0.12 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr14_+_45366518 | 0.12 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr8_+_42195972 | 0.12 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr17_+_48823896 | 0.12 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_+_203734296 | 0.12 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr2_+_234601512 | 0.12 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_171571827 | 0.12 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr2_+_169658928 | 0.11 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr18_+_52385068 | 0.11 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr10_-_14050522 | 0.11 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr15_+_36994210 | 0.11 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr2_+_234602305 | 0.11 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_95963052 | 0.11 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chrX_+_134654540 | 0.11 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr10_+_35484053 | 0.11 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr4_+_75230853 | 0.11 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr8_+_104384616 | 0.11 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr15_-_99057551 | 0.10 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr7_+_111846741 | 0.10 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr4_+_39699664 | 0.10 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chr7_-_83824169 | 0.10 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_215747118 | 0.10 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr2_-_216257849 | 0.10 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr20_+_18447771 | 0.10 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr1_+_162039558 | 0.10 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr1_-_238108575 | 0.10 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr14_+_58765305 | 0.10 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_+_169659121 | 0.09 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr14_+_58765103 | 0.09 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_142374104 | 0.09 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr17_+_26369865 | 0.09 |
ENST00000582037.1
|
NLK
|
nemo-like kinase |
| chr2_+_210518057 | 0.09 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_+_67498396 | 0.09 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr18_-_44775554 | 0.08 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr13_-_41593425 | 0.08 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr4_-_157892055 | 0.08 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr15_-_27018175 | 0.08 |
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr6_+_7389694 | 0.08 |
ENST00000379834.2
|
RIOK1
|
RIO kinase 1 |
| chr2_-_118943930 | 0.08 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr2_+_210444142 | 0.08 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr11_-_31832581 | 0.08 |
ENST00000379111.2
|
PAX6
|
paired box 6 |
| chr8_+_16884740 | 0.08 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr3_+_69811858 | 0.08 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chrX_+_49644470 | 0.08 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr6_+_90272488 | 0.08 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chrX_+_16141667 | 0.08 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chrX_-_54070388 | 0.08 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr11_-_94706705 | 0.08 |
ENST00000279839.6
|
CWC15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr5_-_58295712 | 0.07 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr17_+_26833250 | 0.07 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr18_-_51751132 | 0.07 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr14_-_23652849 | 0.07 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr14_-_90097910 | 0.07 |
ENST00000550332.2
|
RP11-944C7.1
|
Protein LOC100506792 |
| chr6_-_138866823 | 0.07 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr14_+_61788429 | 0.07 |
ENST00000332981.5
|
PRKCH
|
protein kinase C, eta |
| chr12_-_6483969 | 0.07 |
ENST00000396966.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr14_-_37051798 | 0.07 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr12_-_89746264 | 0.07 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr6_-_9933500 | 0.07 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr5_+_137673945 | 0.07 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr3_-_47023455 | 0.07 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr1_+_101361782 | 0.07 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr3_-_185538849 | 0.06 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr4_+_81118647 | 0.06 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr18_-_51750948 | 0.06 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr8_-_22550691 | 0.06 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr4_-_140098339 | 0.06 |
ENST00000394235.2
|
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr9_+_79634571 | 0.06 |
ENST00000376708.1
|
FOXB2
|
forkhead box B2 |
| chr7_+_26331678 | 0.06 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr1_+_155051305 | 0.06 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr14_+_45366472 | 0.06 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr7_-_135194822 | 0.06 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr12_-_57030096 | 0.06 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr15_+_71228826 | 0.06 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr8_-_109260897 | 0.06 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr3_-_88108192 | 0.06 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr6_-_75953484 | 0.06 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr1_-_228613026 | 0.06 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr3_-_196910477 | 0.06 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_23958402 | 0.06 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr9_-_20382446 | 0.06 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr1_-_196577489 | 0.06 |
ENST00000609185.1
ENST00000451324.2 ENST00000367433.5 ENST00000367431.4 |
KCNT2
|
potassium channel, subfamily T, member 2 |
| chr5_+_61874696 | 0.06 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr5_-_111091948 | 0.06 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr3_-_171489085 | 0.06 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr16_+_23194033 | 0.06 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr5_+_161494770 | 0.06 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr16_+_85204882 | 0.06 |
ENST00000574293.1
|
CTC-786C10.1
|
CTC-786C10.1 |
| chr7_-_22234381 | 0.05 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chrX_+_153769446 | 0.05 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr11_+_34999328 | 0.05 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr22_+_38201114 | 0.05 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr6_+_33168189 | 0.05 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr6_-_86352642 | 0.05 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr5_+_61874562 | 0.05 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr16_+_89724188 | 0.05 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr5_-_95550754 | 0.05 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr3_-_88108212 | 0.05 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr6_-_18122843 | 0.05 |
ENST00000340650.3
|
NHLRC1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr1_-_108231101 | 0.05 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr5_+_180682720 | 0.05 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr6_-_86352982 | 0.05 |
ENST00000369622.3
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr9_+_72658490 | 0.05 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr2_+_210517895 | 0.05 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chrX_-_124097620 | 0.05 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr3_-_196910721 | 0.05 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_-_143767881 | 0.05 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr15_+_90544532 | 0.05 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr5_-_124080203 | 0.05 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr9_+_135457530 | 0.05 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr8_+_104310661 | 0.05 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chrX_-_151619746 | 0.05 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr19_+_19496728 | 0.05 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr22_+_45714301 | 0.05 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr6_+_31514622 | 0.05 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr11_-_47206965 | 0.05 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr2_+_166326157 | 0.05 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr19_+_13135386 | 0.05 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr17_+_22022437 | 0.05 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr14_+_93357749 | 0.05 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr10_-_17171785 | 0.04 |
ENST00000377823.1
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr12_-_57037284 | 0.04 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr5_+_140710061 | 0.04 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr1_+_82266053 | 0.04 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr1_+_202385953 | 0.04 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_+_26251835 | 0.04 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr17_+_48823975 | 0.04 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr3_-_112564797 | 0.04 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr8_-_22550815 | 0.04 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr17_+_67498295 | 0.04 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chrX_+_14547632 | 0.04 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr2_-_136743039 | 0.04 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr20_+_9146969 | 0.04 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr7_-_37026108 | 0.04 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_+_19496624 | 0.04 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr9_+_100745615 | 0.04 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr6_-_134499037 | 0.04 |
ENST00000528577.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_-_17533838 | 0.04 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr15_-_63450192 | 0.03 |
ENST00000411926.1
|
RPS27L
|
ribosomal protein S27-like |
| chr20_+_43935474 | 0.03 |
ENST00000372743.1
ENST00000372741.3 ENST00000343694.3 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr11_+_35684288 | 0.03 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chrX_-_24045303 | 0.03 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr12_-_89918522 | 0.03 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr6_-_31514333 | 0.03 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr2_-_190044480 | 0.03 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_-_176733897 | 0.03 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr12_+_56137064 | 0.03 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr5_-_139422654 | 0.03 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chr6_+_44194762 | 0.03 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr5_+_179921344 | 0.03 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr15_+_38544476 | 0.03 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr8_+_85095553 | 0.03 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr4_-_87278857 | 0.03 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_+_88054530 | 0.03 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr1_+_104293028 | 0.03 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr3_+_182511266 | 0.03 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr5_+_50679506 | 0.03 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr18_-_56985776 | 0.03 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr11_+_64009072 | 0.03 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr6_-_94129244 | 0.03 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chrX_-_83442915 | 0.03 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr2_-_136743436 | 0.03 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr1_+_24646002 | 0.03 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr6_+_45296391 | 0.03 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr8_+_42196000 | 0.03 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr6_+_21593972 | 0.03 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr11_-_46142948 | 0.03 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr10_-_31288398 | 0.03 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.0 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |