Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
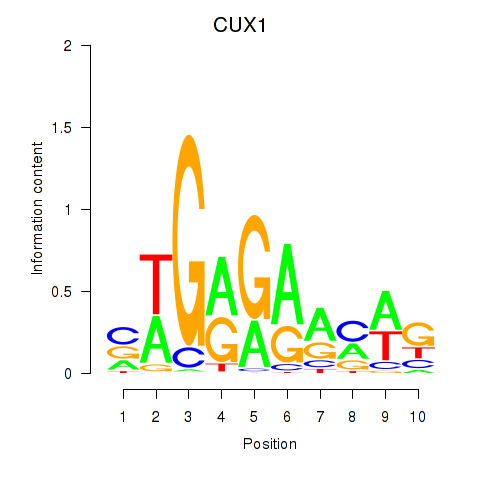
Results for CUX1
Z-value: 1.19
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.5 | cut like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX1 | hg19_v2_chr7_+_101917407_101917445 | 0.36 | 4.8e-01 | Click! |
Activity profile of CUX1 motif
Sorted Z-values of CUX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_7072354 | 0.94 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr9_-_34662651 | 0.83 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr14_+_32476072 | 0.73 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr19_-_10628098 | 0.50 |
ENST00000590601.1
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr5_-_154230130 | 0.47 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr1_-_153513170 | 0.46 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr15_+_78632666 | 0.45 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr19_-_49149553 | 0.44 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr14_+_38065052 | 0.41 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr20_-_62203808 | 0.38 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr8_-_121824374 | 0.38 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr12_-_49581152 | 0.37 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_-_87039662 | 0.37 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr2_-_113594279 | 0.37 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr4_-_155533787 | 0.36 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr12_-_7245080 | 0.33 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr9_-_107754034 | 0.33 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr7_-_99716914 | 0.33 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_161185032 | 0.32 |
ENST00000367992.3
ENST00000289902.1 |
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr14_-_21270995 | 0.30 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr7_-_130597935 | 0.29 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr22_-_30198075 | 0.29 |
ENST00000411532.1
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr5_-_176057518 | 0.29 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr17_-_42906965 | 0.29 |
ENST00000586267.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr11_-_72852320 | 0.28 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr12_-_7245018 | 0.28 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr3_+_142842128 | 0.28 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr4_-_111120334 | 0.28 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr19_-_54604083 | 0.27 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr15_+_91418918 | 0.26 |
ENST00000560824.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_+_114522049 | 0.26 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr3_+_187930429 | 0.26 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr19_-_10213335 | 0.24 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr3_-_142720267 | 0.24 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr8_-_110620284 | 0.24 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr11_+_35222629 | 0.24 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr16_+_2533020 | 0.24 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr19_+_5681011 | 0.24 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr4_+_175204865 | 0.24 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr2_-_218770168 | 0.24 |
ENST00000413554.1
|
TNS1
|
tensin 1 |
| chr14_-_75389925 | 0.23 |
ENST00000556776.1
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr1_-_38325256 | 0.23 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr7_-_127672146 | 0.22 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr11_-_4414880 | 0.22 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr6_-_29600559 | 0.22 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr17_+_77019030 | 0.22 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_-_33182616 | 0.22 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr1_+_180875711 | 0.22 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr6_-_41673552 | 0.21 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr2_-_56150910 | 0.21 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr16_-_30042580 | 0.21 |
ENST00000380495.4
|
FAM57B
|
family with sequence similarity 57, member B |
| chr16_+_2213530 | 0.21 |
ENST00000567645.1
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr7_+_6522922 | 0.20 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr1_-_19615744 | 0.20 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr1_+_155583012 | 0.20 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr8_+_104384616 | 0.20 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr11_-_117688216 | 0.20 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr5_+_149980622 | 0.20 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr2_+_7118755 | 0.20 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr10_+_5238793 | 0.20 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr1_-_235098935 | 0.20 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr3_-_129279894 | 0.20 |
ENST00000506979.1
|
PLXND1
|
plexin D1 |
| chr3_-_165555200 | 0.19 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr19_-_45735138 | 0.19 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr14_-_51863853 | 0.19 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr3_+_187420101 | 0.19 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr11_+_76777979 | 0.19 |
ENST00000531028.1
ENST00000278559.3 ENST00000527066.1 ENST00000529629.1 |
CAPN5
|
calpain 5 |
| chr6_-_31107127 | 0.19 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr17_-_2239729 | 0.19 |
ENST00000576112.2
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr17_+_78397320 | 0.18 |
ENST00000521634.1
ENST00000572886.1 |
ENDOV
|
endonuclease V |
| chr2_+_169923504 | 0.18 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr19_+_54496132 | 0.18 |
ENST00000346968.2
|
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr14_-_24732738 | 0.18 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr19_+_4229495 | 0.18 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr12_+_48499883 | 0.18 |
ENST00000546755.1
ENST00000549366.1 ENST00000552792.1 |
PFKM
|
phosphofructokinase, muscle |
| chr1_-_155948890 | 0.18 |
ENST00000471589.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_+_32812568 | 0.18 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr5_-_53115506 | 0.18 |
ENST00000511953.1
ENST00000504552.1 |
CTD-2081C10.1
|
CTD-2081C10.1 |
| chr12_+_58166726 | 0.18 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr1_-_207206092 | 0.18 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr17_-_42441204 | 0.17 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr5_+_140602904 | 0.17 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr17_+_77018896 | 0.17 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_-_2944184 | 0.17 |
ENST00000337508.4
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr9_+_33795533 | 0.17 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr19_+_10197463 | 0.17 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr10_-_14613968 | 0.17 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr16_-_85784634 | 0.17 |
ENST00000284245.4
ENST00000602914.1 |
C16orf74
|
chromosome 16 open reading frame 74 |
| chr5_-_74348371 | 0.17 |
ENST00000503568.1
|
RP11-229C3.2
|
RP11-229C3.2 |
| chr8_+_22424551 | 0.17 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_119379719 | 0.17 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr3_-_47018219 | 0.17 |
ENST00000292314.2
ENST00000546280.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr22_-_36903069 | 0.17 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr1_-_151345159 | 0.16 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr5_-_132166303 | 0.16 |
ENST00000440118.1
|
SHROOM1
|
shroom family member 1 |
| chr11_+_45943169 | 0.16 |
ENST00000529052.1
ENST00000531526.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr2_+_242312264 | 0.16 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr6_+_32811885 | 0.16 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_-_32812420 | 0.16 |
ENST00000374881.2
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr3_-_114790179 | 0.16 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_32820529 | 0.16 |
ENST00000425148.2
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr9_-_115983641 | 0.16 |
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr3_-_49933506 | 0.16 |
ENST00000440292.1
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr19_-_41220957 | 0.16 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr16_-_1494490 | 0.16 |
ENST00000389176.3
ENST00000409671.1 |
CCDC154
|
coiled-coil domain containing 154 |
| chr17_+_14277419 | 0.16 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chr1_+_36621529 | 0.15 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr5_-_131826457 | 0.15 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr16_-_56223480 | 0.15 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr2_-_31637560 | 0.15 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr9_+_127213423 | 0.15 |
ENST00000334810.1
|
GPR144
|
G protein-coupled receptor 144 |
| chr14_+_24702073 | 0.15 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr22_-_41940404 | 0.15 |
ENST00000355209.4
ENST00000337566.5 ENST00000396504.2 ENST00000407461.1 |
POLR3H
|
polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) |
| chr2_+_132233664 | 0.15 |
ENST00000321253.6
|
TUBA3D
|
tubulin, alpha 3d |
| chr7_-_75443118 | 0.15 |
ENST00000222902.2
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr17_+_62075703 | 0.15 |
ENST00000577953.1
ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72
|
chromosome 17 open reading frame 72 |
| chr17_+_6900201 | 0.15 |
ENST00000480801.1
|
ALOX12
|
arachidonate 12-lipoxygenase |
| chr17_+_1665345 | 0.15 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr4_-_129491686 | 0.14 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr3_-_194207388 | 0.14 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr17_+_1665253 | 0.14 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_+_70036164 | 0.14 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr19_-_55658687 | 0.14 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr14_+_24779376 | 0.14 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr10_-_14050522 | 0.14 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr1_+_48688357 | 0.14 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr7_-_99527243 | 0.14 |
ENST00000312891.2
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa |
| chr3_+_69985792 | 0.14 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr19_-_10450328 | 0.14 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr19_-_58485895 | 0.14 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr19_-_1650666 | 0.13 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr19_+_35645817 | 0.13 |
ENST00000423817.3
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr14_-_70546897 | 0.13 |
ENST00000394330.2
ENST00000533541.1 ENST00000216568.7 |
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr6_-_53013620 | 0.13 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr1_+_203096831 | 0.13 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr10_-_51958906 | 0.13 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr2_+_27719697 | 0.13 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr5_+_150020240 | 0.13 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr15_-_90294523 | 0.13 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr9_-_130667592 | 0.13 |
ENST00000447681.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr16_-_89768097 | 0.13 |
ENST00000289805.5
ENST00000335360.7 |
SPATA2L
|
spermatogenesis associated 2-like |
| chr19_-_12941469 | 0.13 |
ENST00000586969.1
ENST00000589681.1 ENST00000585384.1 ENST00000589808.1 |
RTBDN
|
retbindin |
| chr8_-_74005349 | 0.13 |
ENST00000297354.6
|
SBSPON
|
somatomedin B and thrombospondin, type 1 domain containing |
| chr22_+_27068704 | 0.13 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chrX_+_10126488 | 0.13 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr18_+_72166564 | 0.13 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr19_+_45312310 | 0.13 |
ENST00000589651.1
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr9_-_113800705 | 0.13 |
ENST00000441240.1
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr16_+_24857552 | 0.13 |
ENST00000568579.1
ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr16_+_24857309 | 0.13 |
ENST00000565769.1
ENST00000449109.2 ENST00000424767.2 ENST00000545376.1 ENST00000569520.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr19_+_55795493 | 0.13 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr7_+_134430212 | 0.13 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr1_-_67142710 | 0.13 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr19_-_50381606 | 0.13 |
ENST00000391830.1
|
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr15_-_41522889 | 0.12 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr16_+_67571351 | 0.12 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr17_-_18950310 | 0.12 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr11_+_73675873 | 0.12 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr19_-_44258733 | 0.12 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr22_+_51176624 | 0.12 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr22_-_50451088 | 0.12 |
ENST00000389983.2
|
IL17REL
|
interleukin 17 receptor E-like |
| chr6_+_32811861 | 0.12 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr19_-_55658281 | 0.12 |
ENST00000585321.2
ENST00000587465.2 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr17_+_65374075 | 0.12 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr12_-_111358372 | 0.12 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr19_-_11529225 | 0.12 |
ENST00000567431.1
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr12_-_56753858 | 0.12 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr14_-_73924954 | 0.12 |
ENST00000555307.1
ENST00000554818.1 |
NUMB
|
numb homolog (Drosophila) |
| chr1_-_212588157 | 0.12 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr1_+_113263041 | 0.12 |
ENST00000369630.3
|
FAM19A3
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
| chr6_-_41703952 | 0.12 |
ENST00000358871.2
ENST00000403298.4 |
TFEB
|
transcription factor EB |
| chr16_+_27413483 | 0.12 |
ENST00000337929.3
ENST00000564089.1 |
IL21R
|
interleukin 21 receptor |
| chr17_-_56605341 | 0.12 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr9_+_115983808 | 0.12 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr16_+_30935896 | 0.12 |
ENST00000562319.1
ENST00000380310.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr17_+_39868577 | 0.12 |
ENST00000329402.3
|
GAST
|
gastrin |
| chr17_+_45908974 | 0.12 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr2_-_73869508 | 0.12 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr1_-_45288932 | 0.12 |
ENST00000438067.1
|
PTCH2
|
patched 2 |
| chr17_-_38928414 | 0.11 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr17_+_75315654 | 0.11 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr20_+_44034676 | 0.11 |
ENST00000372723.3
ENST00000372722.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_62186498 | 0.11 |
ENST00000278282.2
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr1_-_115632035 | 0.11 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr3_-_48542197 | 0.11 |
ENST00000442747.1
ENST00000444115.1 |
SHISA5
|
shisa family member 5 |
| chr1_+_26605618 | 0.11 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_+_45417812 | 0.11 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr12_-_58287735 | 0.11 |
ENST00000548955.1
|
RP11-620J15.2
|
RP11-620J15.2 |
| chr19_-_1095330 | 0.11 |
ENST00000586746.1
|
POLR2E
|
polymerase (RNA) II (DNA directed) polypeptide E, 25kDa |
| chr3_-_134093738 | 0.11 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr5_+_150157444 | 0.11 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr12_-_48499826 | 0.11 |
ENST00000551798.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr1_+_206730484 | 0.11 |
ENST00000304534.8
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr2_-_128784846 | 0.11 |
ENST00000259235.3
ENST00000357702.5 ENST00000424298.1 |
SAP130
|
Sin3A-associated protein, 130kDa |
| chr8_+_107282368 | 0.11 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr7_-_102252038 | 0.11 |
ENST00000461209.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr5_-_135290705 | 0.11 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_138730817 | 0.11 |
ENST00000434752.2
|
PROB1
|
proline-rich basic protein 1 |
| chr1_-_155177677 | 0.11 |
ENST00000368378.3
ENST00000541990.1 ENST00000457183.2 |
THBS3
|
thrombospondin 3 |
| chr5_+_175288631 | 0.11 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001812 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type I hypersensitivity(GO:0001812) positive regulation of type II hypersensitivity(GO:0002894) positive regulation of mast cell cytokine production(GO:0032765) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.4 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.3 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.7 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.1 | 0.5 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0100012 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.1 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0003289 | septum primum development(GO:0003284) septum secundum development(GO:0003285) atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:1990748 | detoxification(GO:0098754) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0036006 | negative regulation of protein import into nucleus, translocation(GO:0033159) response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.0 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.2 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |