Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
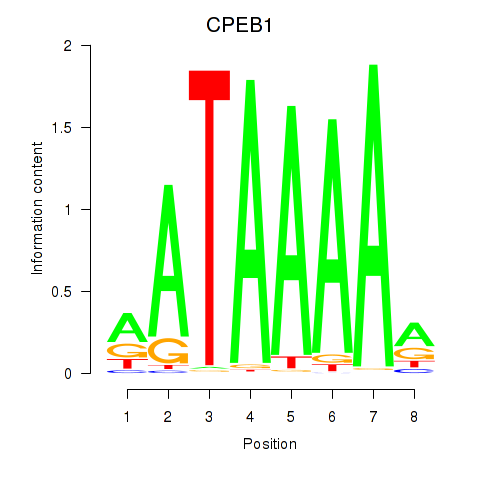
Results for CPEB1
Z-value: 1.03
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83240783_83240846 | -0.84 | 3.5e-02 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_148779106 | 1.03 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr2_+_162101247 | 1.02 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr1_-_65533390 | 0.96 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr1_+_95616933 | 0.88 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr3_-_114035026 | 0.75 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr22_-_41258074 | 0.75 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr3_-_148939598 | 0.64 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr11_+_28724129 | 0.58 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr10_+_94590910 | 0.58 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_72058130 | 0.55 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr8_-_90993869 | 0.53 |
ENST00000517772.1
|
NBN
|
nibrin |
| chrX_-_20237059 | 0.50 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr7_-_33080506 | 0.49 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr6_+_26104104 | 0.49 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr7_-_27205136 | 0.48 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr13_+_28519343 | 0.47 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr5_+_32788945 | 0.47 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr5_+_139505520 | 0.47 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr9_-_69229650 | 0.46 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr8_+_71485681 | 0.46 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr12_-_91546926 | 0.45 |
ENST00000550758.1
|
DCN
|
decorin |
| chr9_+_22646189 | 0.42 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr1_-_200379180 | 0.40 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr1_+_205682497 | 0.39 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr18_-_68004529 | 0.38 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr1_+_116654376 | 0.38 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr2_-_148778323 | 0.38 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chrX_-_138914394 | 0.38 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr6_+_27114861 | 0.37 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr2_+_145780739 | 0.37 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chrX_+_72783026 | 0.36 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr10_-_21806759 | 0.36 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr5_+_56471592 | 0.36 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr6_+_119215308 | 0.35 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr9_-_70465758 | 0.35 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr9_-_16253112 | 0.35 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr13_-_44735393 | 0.35 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr2_-_151344172 | 0.35 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr2_+_176987088 | 0.34 |
ENST00000249499.6
|
HOXD9
|
homeobox D9 |
| chr3_-_57233966 | 0.34 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr2_-_87248975 | 0.34 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr14_+_73563735 | 0.33 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr4_-_65275162 | 0.33 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr6_-_10115007 | 0.32 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr2_+_64073187 | 0.32 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr1_+_209848749 | 0.32 |
ENST00000367029.4
|
G0S2
|
G0/G1switch 2 |
| chr10_-_106240032 | 0.32 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr1_+_61869748 | 0.32 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chrM_+_5824 | 0.30 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr17_+_39846114 | 0.30 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr8_+_38831683 | 0.30 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr9_+_42671887 | 0.30 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chrX_-_77395186 | 0.29 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr2_+_67624430 | 0.29 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr6_-_32083106 | 0.29 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chr6_+_111195973 | 0.28 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr7_-_83824449 | 0.28 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_+_7811992 | 0.28 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr2_-_206950781 | 0.28 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr2_-_148778258 | 0.28 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr16_+_2820912 | 0.27 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr8_-_25281747 | 0.27 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr19_+_36706024 | 0.27 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chr8_+_77596014 | 0.27 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr10_+_114709999 | 0.27 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr14_+_102228123 | 0.26 |
ENST00000422945.2
ENST00000554442.1 ENST00000556260.2 ENST00000328724.5 ENST00000557268.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_84629976 | 0.26 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_28283069 | 0.26 |
ENST00000466830.1
ENST00000423894.1 |
CMC1
|
C-x(9)-C motif containing 1 |
| chr10_+_99627889 | 0.25 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr8_-_57123815 | 0.25 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr19_-_35992780 | 0.25 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr11_+_34642656 | 0.25 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr1_+_84630574 | 0.25 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr18_+_29769978 | 0.25 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr3_+_160394940 | 0.25 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr9_+_97562440 | 0.25 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr4_+_174089904 | 0.25 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chrX_+_123095890 | 0.24 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr2_+_192543694 | 0.24 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_-_38574169 | 0.24 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr11_+_77300669 | 0.24 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr1_-_108231101 | 0.23 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr5_-_111091948 | 0.23 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr4_-_48782259 | 0.23 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr21_-_35340759 | 0.23 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr17_+_7533439 | 0.23 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr14_+_56078695 | 0.23 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr16_-_3422283 | 0.23 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr7_-_14029515 | 0.23 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr19_-_57967854 | 0.22 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr17_+_45286387 | 0.22 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_+_145780725 | 0.22 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr4_-_105416039 | 0.22 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr3_-_93747425 | 0.22 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr17_+_7461613 | 0.21 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr5_-_76935513 | 0.21 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr10_-_52008313 | 0.21 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr6_-_121655593 | 0.21 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr7_-_14029283 | 0.21 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr15_-_56209306 | 0.21 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr7_+_142374104 | 0.20 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr3_+_142342228 | 0.20 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr5_+_173316341 | 0.20 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_+_164528616 | 0.20 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr11_+_17281900 | 0.20 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr2_-_145277569 | 0.20 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_151739131 | 0.19 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr1_+_82266053 | 0.19 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr1_-_103574024 | 0.19 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr6_+_117002339 | 0.19 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr14_+_38091270 | 0.19 |
ENST00000553443.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr2_-_145277640 | 0.18 |
ENST00000539609.3
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_169626104 | 0.18 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr1_-_11918988 | 0.18 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr14_-_35344093 | 0.18 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr4_-_70725856 | 0.18 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chrM_+_14741 | 0.18 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr8_-_95274536 | 0.18 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chrX_+_123095546 | 0.18 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr1_+_84630352 | 0.18 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_-_71869642 | 0.17 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr7_-_75401513 | 0.17 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr12_-_76817036 | 0.17 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr15_-_76304731 | 0.17 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr1_+_212475148 | 0.17 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr8_+_94752349 | 0.17 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr16_-_12062333 | 0.17 |
ENST00000597717.1
|
AC007216.2
|
Uncharacterized protein |
| chr1_+_169077172 | 0.17 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr6_-_45544507 | 0.17 |
ENST00000563807.1
|
RP1-166H4.2
|
RP1-166H4.2 |
| chr6_+_62284008 | 0.17 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr15_-_55790515 | 0.17 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chrX_+_123095860 | 0.17 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr9_-_20622478 | 0.17 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr15_+_40886199 | 0.17 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chrX_+_73164167 | 0.17 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr14_+_39703112 | 0.16 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr16_+_53133070 | 0.16 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_+_74301880 | 0.16 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr8_-_108510224 | 0.16 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr20_+_9049303 | 0.16 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr1_+_167298281 | 0.15 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr3_-_196910477 | 0.15 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr9_+_74920335 | 0.15 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr18_+_29171689 | 0.15 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr12_-_92536433 | 0.15 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chrX_+_133733457 | 0.15 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr3_-_69129501 | 0.15 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr4_-_69111401 | 0.15 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr2_-_55646412 | 0.15 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr12_-_54071181 | 0.15 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr18_-_33709268 | 0.15 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chrX_-_46187069 | 0.14 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr3_+_186501336 | 0.14 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr17_-_46806540 | 0.14 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr9_+_82188077 | 0.14 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chrM_-_14670 | 0.14 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr17_-_53800217 | 0.14 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chr14_-_102552659 | 0.14 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr18_-_60985914 | 0.14 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr3_-_155394152 | 0.14 |
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr6_-_121655552 | 0.14 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr12_-_53228079 | 0.13 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr7_+_115862858 | 0.13 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr1_+_84630053 | 0.13 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_-_92413727 | 0.13 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr1_-_115259337 | 0.13 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr14_-_62217779 | 0.13 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr12_+_41221975 | 0.13 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr3_+_107241783 | 0.13 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_27114577 | 0.13 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr3_-_108248169 | 0.13 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr13_+_49684445 | 0.13 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr7_+_90012986 | 0.13 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr2_-_61389240 | 0.13 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr6_-_64029879 | 0.13 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chrX_+_78003204 | 0.13 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chrX_+_73164149 | 0.13 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr2_+_145780767 | 0.13 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr14_+_79745682 | 0.13 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr7_+_129906660 | 0.13 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr14_-_64846033 | 0.12 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr2_-_152118352 | 0.12 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chrX_-_20236970 | 0.12 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr14_+_39703084 | 0.12 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr16_+_16472912 | 0.12 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr12_+_41221794 | 0.12 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr5_+_110073853 | 0.12 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr1_+_154966058 | 0.12 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr3_-_117716418 | 0.12 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr10_+_114710211 | 0.12 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr11_-_61196858 | 0.12 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_+_181845843 | 0.12 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr12_+_28410128 | 0.12 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr18_+_61575200 | 0.12 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr2_+_105050794 | 0.12 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr1_+_18957500 | 0.12 |
ENST00000375375.3
|
PAX7
|
paired box 7 |
| chr6_+_127898312 | 0.12 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr7_-_16921601 | 0.12 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr12_-_52845910 | 0.12 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr4_+_144303093 | 0.11 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.4 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0070668 | regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0060345 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.2 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.3 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:2000510 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:1900194 | female pronucleus assembly(GO:0035038) negative regulation of oocyte maturation(GO:1900194) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 1.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.3 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |