Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
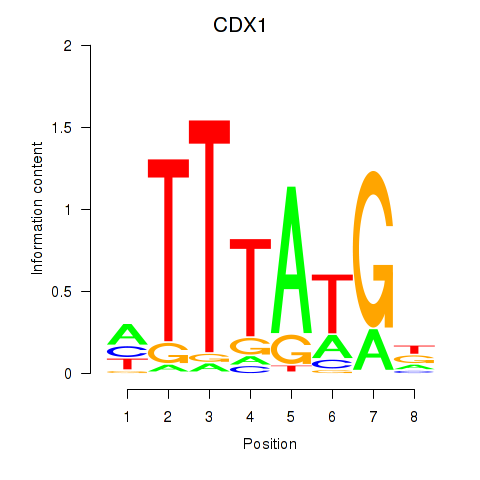
Results for CDX1
Z-value: 0.50
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | caudal type homeobox 1 |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_33080506 | 0.50 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_-_35013217 | 0.48 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr2_+_172309634 | 0.39 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr7_-_92777606 | 0.37 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr14_-_54955721 | 0.36 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr2_+_161993465 | 0.34 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr16_+_56672571 | 0.33 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr3_-_148939598 | 0.31 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr21_-_35773370 | 0.28 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr8_-_125551278 | 0.28 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr3_-_96337000 | 0.27 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr9_-_69229650 | 0.27 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr2_+_161993412 | 0.25 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_193075180 | 0.25 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr15_+_49715449 | 0.25 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr14_+_58797974 | 0.24 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr19_+_52076425 | 0.24 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chrX_-_119693745 | 0.24 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr8_+_86121448 | 0.24 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chrX_-_80377118 | 0.23 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr5_-_150138061 | 0.23 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr12_+_20963647 | 0.22 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr5_+_118691706 | 0.22 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chrY_+_14958970 | 0.22 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr8_-_42358742 | 0.22 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr8_-_123706338 | 0.22 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr18_-_68004529 | 0.22 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr4_+_26324474 | 0.21 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chrX_-_45629661 | 0.21 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr4_+_74606223 | 0.21 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr2_-_151344172 | 0.21 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr13_+_28519343 | 0.20 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr14_+_102276209 | 0.20 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr5_+_75904950 | 0.20 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr21_+_17214724 | 0.20 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr19_-_23578220 | 0.20 |
ENST00000595533.1
ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91
|
zinc finger protein 91 |
| chr19_+_46367518 | 0.20 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr2_+_192543694 | 0.19 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr7_+_79765071 | 0.19 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr10_-_69597828 | 0.19 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr12_-_10324716 | 0.19 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr2_+_242289502 | 0.19 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chrM_+_14741 | 0.19 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr10_-_36813162 | 0.18 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr1_-_54411240 | 0.18 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_-_76461249 | 0.18 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_150254936 | 0.18 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr14_+_67831576 | 0.18 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr17_+_65713925 | 0.17 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr16_+_56691606 | 0.17 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr16_+_53241854 | 0.17 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr12_-_90049878 | 0.17 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_+_20963632 | 0.17 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_100485143 | 0.17 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr12_+_16500037 | 0.17 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr13_+_73302047 | 0.17 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr11_-_85780853 | 0.17 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_68299130 | 0.16 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr1_+_212475148 | 0.16 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr14_+_65878650 | 0.16 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr12_-_118796971 | 0.16 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr5_-_39203093 | 0.16 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr8_+_76452097 | 0.16 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr7_+_120629653 | 0.16 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_19748640 | 0.16 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr5_-_79946820 | 0.16 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr7_-_16840820 | 0.15 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr10_-_38146965 | 0.15 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr11_-_10530723 | 0.15 |
ENST00000536684.1
|
MTRNR2L8
|
MT-RNR2-like 8 |
| chr6_-_110011718 | 0.15 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr10_+_112327425 | 0.15 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr5_+_56471592 | 0.15 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr6_+_88299833 | 0.15 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr21_+_25801041 | 0.15 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr9_-_3469181 | 0.15 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr1_+_178694300 | 0.15 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_+_57358750 | 0.15 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr17_+_22022437 | 0.15 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr1_-_54411255 | 0.14 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr8_+_107738240 | 0.14 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr18_+_20513782 | 0.14 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr19_+_38880695 | 0.14 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_115177178 | 0.13 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr4_+_156824840 | 0.13 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr13_+_53030107 | 0.13 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr5_-_96518907 | 0.13 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr1_+_89246647 | 0.13 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr15_+_45879964 | 0.13 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr2_-_44550441 | 0.13 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr5_+_75904918 | 0.13 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr15_+_49715293 | 0.13 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr2_+_46926048 | 0.13 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr11_+_107992243 | 0.13 |
ENST00000265838.4
ENST00000299355.6 |
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr2_+_192543153 | 0.13 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_28410128 | 0.13 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr2_+_196521458 | 0.13 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr18_-_53303123 | 0.13 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr2_-_37544209 | 0.12 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr14_+_75179840 | 0.12 |
ENST00000554590.1
ENST00000341162.4 ENST00000534938.2 ENST00000553615.1 |
FCF1
|
FCF1 rRNA-processing protein |
| chr14_+_53196872 | 0.12 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr7_+_105172532 | 0.12 |
ENST00000257700.2
|
RINT1
|
RAD50 interactor 1 |
| chr7_+_141463897 | 0.12 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr11_-_2158507 | 0.12 |
ENST00000381392.1
ENST00000381395.1 ENST00000418738.2 |
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr7_-_54826920 | 0.12 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr4_-_175443943 | 0.12 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr8_+_16884740 | 0.12 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr4_+_154073469 | 0.12 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr15_+_77713222 | 0.12 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chrX_-_122756660 | 0.12 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr5_+_64920543 | 0.12 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr5_+_118691008 | 0.11 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr12_-_90049828 | 0.11 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_+_117220016 | 0.11 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr6_-_134373732 | 0.11 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr15_-_55700457 | 0.11 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr8_+_103876528 | 0.11 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr5_-_95550754 | 0.11 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr14_-_67826486 | 0.11 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr14_-_64846033 | 0.11 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr14_+_24407940 | 0.11 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr3_+_190333097 | 0.10 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr17_+_68164752 | 0.10 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr7_+_6714599 | 0.10 |
ENST00000328239.7
ENST00000542006.1 |
AC073343.1
|
Uncharacterized protein |
| chr2_+_192542879 | 0.10 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_-_48943706 | 0.10 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr10_-_69597915 | 0.10 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chrX_-_24045303 | 0.10 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr18_-_57027194 | 0.10 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr15_-_55700522 | 0.10 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr14_+_96722539 | 0.10 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr14_+_56127960 | 0.10 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chrX_+_591524 | 0.09 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr8_+_107738343 | 0.09 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr1_+_84629976 | 0.09 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_+_86748898 | 0.09 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr7_-_130353553 | 0.09 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr1_-_28384598 | 0.09 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr4_-_85771168 | 0.09 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr18_+_47901408 | 0.09 |
ENST00000398452.2
ENST00000417656.2 ENST00000488454.1 ENST00000494518.1 |
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr9_-_21335240 | 0.09 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr1_+_81771806 | 0.09 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr14_+_58894706 | 0.09 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr2_+_32502952 | 0.09 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chrM_-_14670 | 0.09 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr8_-_103876383 | 0.09 |
ENST00000347770.4
|
AZIN1
|
antizyme inhibitor 1 |
| chr12_+_9144626 | 0.09 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr12_+_59989918 | 0.09 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr4_+_154622652 | 0.08 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr15_+_55700741 | 0.08 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr6_-_47009996 | 0.08 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr8_-_117886955 | 0.08 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr12_-_70093162 | 0.08 |
ENST00000551160.1
|
BEST3
|
bestrophin 3 |
| chr5_-_133510456 | 0.08 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr7_-_29152509 | 0.08 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr18_-_61329118 | 0.08 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr12_+_72079842 | 0.08 |
ENST00000266673.5
ENST00000550524.1 |
TMEM19
|
transmembrane protein 19 |
| chr3_+_182983090 | 0.08 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr8_+_19536083 | 0.08 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr13_+_78109884 | 0.08 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr1_+_196743943 | 0.08 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr8_+_93895865 | 0.08 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr6_+_63921351 | 0.08 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr1_-_86848760 | 0.08 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr10_-_95360983 | 0.08 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr1_+_84630574 | 0.07 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_+_102201509 | 0.07 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr9_-_21335356 | 0.07 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr5_-_75008244 | 0.07 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr2_-_165424973 | 0.07 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr13_-_49975632 | 0.07 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr17_+_57233087 | 0.07 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr1_+_161736072 | 0.07 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr4_+_175205038 | 0.07 |
ENST00000457424.2
ENST00000514712.1 |
CEP44
|
centrosomal protein 44kDa |
| chr13_+_49684445 | 0.07 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr17_-_57232596 | 0.07 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr6_+_143999072 | 0.07 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr14_+_61201445 | 0.07 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr17_+_65375082 | 0.07 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr4_-_100485183 | 0.07 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr7_+_77469439 | 0.07 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr9_+_125795788 | 0.07 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr5_+_68530697 | 0.07 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr7_+_90012986 | 0.07 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr17_-_29641104 | 0.07 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr2_+_89901292 | 0.07 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr7_+_23210760 | 0.07 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr3_+_107364683 | 0.07 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr5_+_174905398 | 0.06 |
ENST00000321442.5
|
SFXN1
|
sideroflexin 1 |
| chr5_-_76916396 | 0.06 |
ENST00000509971.1
|
WDR41
|
WD repeat domain 41 |
| chr14_+_37641012 | 0.06 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chrX_+_13707235 | 0.06 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr1_+_84630053 | 0.06 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_+_19389814 | 0.06 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr16_-_28192360 | 0.06 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr15_+_42066888 | 0.06 |
ENST00000510535.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr4_+_86749045 | 0.06 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr18_-_24443151 | 0.06 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chr12_+_97306295 | 0.06 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr11_+_28129795 | 0.06 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr5_+_108083517 | 0.06 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.2 | GO:0061565 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.5 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.3 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.0 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |