Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
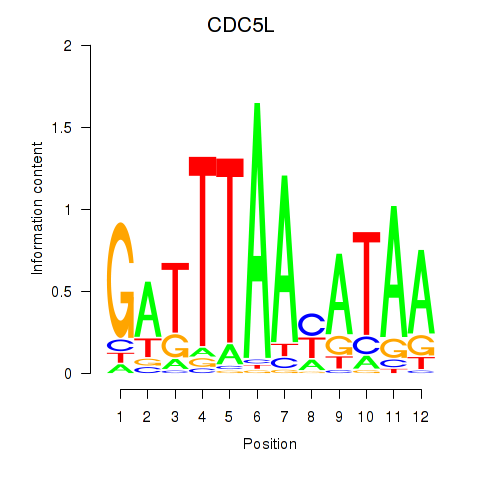
Results for CDC5L
Z-value: 1.50
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | 0.80 | 5.4e-02 | Click! |
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_91152303 | 5.41 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr4_+_155484155 | 1.32 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr3_-_122283424 | 1.19 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr15_-_42076229 | 1.19 |
ENST00000597767.1
|
AC073657.1
|
Uncharacterized protein |
| chr4_+_155484103 | 1.16 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr22_-_40929812 | 0.95 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr2_+_228678550 | 0.90 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr4_+_80584903 | 0.87 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr1_+_196621002 | 0.86 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr18_+_61442629 | 0.83 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr10_-_69597828 | 0.75 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr21_+_37692481 | 0.74 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr1_+_158985457 | 0.74 |
ENST00000567661.1
ENST00000474473.1 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr5_-_133510456 | 0.73 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr4_+_74606223 | 0.72 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr10_-_69597810 | 0.71 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_+_200472779 | 0.70 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chrX_-_11129229 | 0.69 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr1_-_154600421 | 0.68 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr5_-_157161727 | 0.67 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr19_-_54567159 | 0.64 |
ENST00000338372.2
ENST00000376626.1 |
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr2_-_203103185 | 0.64 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr2_-_113542063 | 0.63 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr13_+_78315528 | 0.61 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr18_+_20494078 | 0.61 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr3_+_107364769 | 0.61 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr7_-_102985288 | 0.59 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr13_-_86373536 | 0.58 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr1_+_196621156 | 0.58 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr6_-_76072719 | 0.57 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_-_69597915 | 0.57 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr18_+_61445007 | 0.56 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr3_+_108321623 | 0.55 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr17_+_68164752 | 0.53 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_+_127140956 | 0.53 |
ENST00000608214.1
|
RP11-480C22.1
|
RP11-480C22.1 |
| chr9_-_73029540 | 0.52 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr4_+_164265035 | 0.51 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr20_+_52824367 | 0.50 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr11_-_102651343 | 0.49 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr3_+_57882061 | 0.49 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr1_-_89591749 | 0.47 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr3_+_107364683 | 0.47 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr9_+_35673853 | 0.47 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr8_+_107593198 | 0.46 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr13_-_31038370 | 0.46 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr19_+_51897742 | 0.46 |
ENST00000600765.1
|
CTD-2616J11.14
|
CTD-2616J11.14 |
| chr2_+_238877424 | 0.46 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr14_-_54425475 | 0.45 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr12_-_53601055 | 0.45 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr10_+_126630692 | 0.45 |
ENST00000359653.4
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1 |
| chr14_+_104029362 | 0.43 |
ENST00000495778.1
|
APOPT1
|
apoptogenic 1, mitochondrial |
| chr10_+_27793257 | 0.43 |
ENST00000375802.3
|
RAB18
|
RAB18, member RAS oncogene family |
| chr2_-_180871780 | 0.43 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_+_93645314 | 0.42 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr3_+_122044084 | 0.42 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr11_-_85430088 | 0.42 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr14_+_56127960 | 0.42 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr4_+_119200215 | 0.42 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr14_+_56127989 | 0.42 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_+_78470530 | 0.41 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr6_-_74233480 | 0.41 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr12_-_49463620 | 0.40 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr1_-_247094628 | 0.39 |
ENST00000366508.1
ENST00000326225.3 ENST00000391829.2 |
AHCTF1
|
AT hook containing transcription factor 1 |
| chr21_+_35107346 | 0.39 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr21_+_17214724 | 0.38 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr6_+_64345698 | 0.38 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr7_+_33945132 | 0.38 |
ENST00000436222.1
|
BMPER
|
BMP binding endothelial regulator |
| chrM_+_12331 | 0.38 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr11_-_62609281 | 0.38 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr12_+_79439405 | 0.37 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr10_+_111765562 | 0.37 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr19_+_37178482 | 0.37 |
ENST00000536254.2
|
ZNF567
|
zinc finger protein 567 |
| chr21_+_33784670 | 0.37 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr4_+_130017268 | 0.36 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr5_-_135290651 | 0.36 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr10_-_62493223 | 0.36 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_150039877 | 0.36 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr12_+_27398584 | 0.36 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr18_-_37380230 | 0.35 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr2_+_86116396 | 0.35 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr2_+_210444748 | 0.35 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr6_+_26156551 | 0.35 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr22_+_29168652 | 0.35 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr4_-_76912070 | 0.35 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr13_+_78315348 | 0.34 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr13_+_76378305 | 0.33 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr3_-_107596910 | 0.33 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr2_+_86333340 | 0.33 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr17_+_15604513 | 0.33 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr2_+_196521845 | 0.33 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_+_57881966 | 0.33 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chr7_+_128399002 | 0.33 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr2_+_210518057 | 0.33 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_70820451 | 0.33 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chrX_-_45629661 | 0.32 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr6_+_76330355 | 0.32 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr1_+_179335101 | 0.32 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr2_-_203103281 | 0.32 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr1_+_156308403 | 0.31 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr4_-_185395191 | 0.31 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr3_-_64009658 | 0.31 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr15_+_49715449 | 0.31 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr4_-_57253587 | 0.31 |
ENST00000513376.1
ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr5_+_68860949 | 0.31 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr3_+_25824405 | 0.30 |
ENST00000452098.1
|
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr1_-_209741018 | 0.30 |
ENST00000424696.2
|
RP1-272L16.1
|
RP1-272L16.1 |
| chr2_+_132479948 | 0.30 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr3_+_151531859 | 0.30 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr1_-_150738261 | 0.30 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr12_-_57030096 | 0.30 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr7_-_102985035 | 0.29 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr6_-_56492816 | 0.29 |
ENST00000522360.1
|
DST
|
dystonin |
| chr14_-_38036271 | 0.29 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr5_-_43557791 | 0.29 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr5_+_179135246 | 0.29 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr14_-_53331239 | 0.29 |
ENST00000553663.1
|
FERMT2
|
fermitin family member 2 |
| chr18_-_28682374 | 0.29 |
ENST00000280904.6
|
DSC2
|
desmocollin 2 |
| chr8_-_124279627 | 0.28 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr1_-_65468148 | 0.28 |
ENST00000415842.1
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr19_+_21324863 | 0.28 |
ENST00000598331.1
|
ZNF431
|
zinc finger protein 431 |
| chr14_-_31856397 | 0.28 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr12_+_28605426 | 0.27 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_244515930 | 0.27 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr11_-_31014214 | 0.27 |
ENST00000406071.2
ENST00000339794.5 |
DCDC1
|
doublecortin domain containing 1 |
| chr11_+_73661364 | 0.27 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr8_+_104311059 | 0.27 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chrX_+_23925918 | 0.27 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr5_-_95018660 | 0.26 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr15_-_55562451 | 0.26 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_+_65823092 | 0.26 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr3_+_57882024 | 0.26 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr12_+_1099675 | 0.26 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_+_33683109 | 0.26 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_44550441 | 0.26 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr2_+_234601512 | 0.26 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr16_+_21623392 | 0.26 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr5_+_43033818 | 0.26 |
ENST00000607830.1
|
CTD-2035E11.4
|
CTD-2035E11.4 |
| chr3_+_20081515 | 0.26 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr3_-_149051444 | 0.26 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr3_-_176914238 | 0.26 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_+_118572226 | 0.26 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr3_+_177545563 | 0.26 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr8_-_110988070 | 0.26 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr10_+_35484053 | 0.26 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr2_-_37544209 | 0.26 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr16_-_69418553 | 0.25 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr2_+_44001172 | 0.25 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr19_+_852291 | 0.25 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr9_+_40028620 | 0.25 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr5_+_33440802 | 0.25 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr2_-_191399073 | 0.25 |
ENST00000421038.1
|
TMEM194B
|
transmembrane protein 194B |
| chr12_+_19593515 | 0.25 |
ENST00000360995.4
|
AEBP2
|
AE binding protein 2 |
| chr1_-_6260896 | 0.25 |
ENST00000497965.1
|
RPL22
|
ribosomal protein L22 |
| chr5_-_98262240 | 0.25 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr4_+_170541678 | 0.25 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr12_-_76817036 | 0.25 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr7_+_77469439 | 0.24 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_+_198189921 | 0.24 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr6_-_149969829 | 0.24 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr12_-_54653313 | 0.24 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr2_+_183582774 | 0.24 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr10_+_27793197 | 0.24 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr1_+_150039787 | 0.24 |
ENST00000535106.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr3_+_184534994 | 0.24 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr11_-_85430163 | 0.24 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_78804393 | 0.23 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr5_+_121297650 | 0.23 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr6_+_126221034 | 0.23 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr18_-_10701979 | 0.23 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr6_-_149969871 | 0.23 |
ENST00000335643.8
ENST00000444282.1 |
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr22_+_44761431 | 0.23 |
ENST00000406912.1
|
RP1-32I10.10
|
Uncharacterized protein |
| chr1_+_156338993 | 0.23 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr9_-_20382446 | 0.23 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr6_+_57182400 | 0.23 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr1_+_63249796 | 0.23 |
ENST00000443289.1
ENST00000317868.4 ENST00000371120.3 |
ATG4C
|
autophagy related 4C, cysteine peptidase |
| chrX_-_15683147 | 0.23 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr14_-_60636561 | 0.23 |
ENST00000536410.2
ENST00000216500.5 |
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr4_+_119809984 | 0.23 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr11_+_57308979 | 0.23 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr8_+_92082424 | 0.22 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr20_+_16710606 | 0.22 |
ENST00000377943.5
ENST00000246071.6 |
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B |
| chr6_+_27791862 | 0.22 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr5_-_148929848 | 0.22 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr4_+_48833015 | 0.22 |
ENST00000509122.1
ENST00000509664.1 ENST00000505922.2 ENST00000514981.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr16_-_69419473 | 0.22 |
ENST00000566750.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr8_+_95731904 | 0.22 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr13_+_50589390 | 0.22 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr4_-_141348999 | 0.22 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr3_-_176914191 | 0.22 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_+_210517895 | 0.22 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_66458072 | 0.22 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr2_+_70121075 | 0.21 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr8_-_62602327 | 0.21 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr2_+_196521903 | 0.21 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr16_-_3422283 | 0.21 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr6_+_121756809 | 0.21 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr4_-_159094194 | 0.21 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr18_-_11148587 | 0.21 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr10_+_69865866 | 0.21 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr6_+_136172820 | 0.21 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 1.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.3 | 0.9 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.7 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 2.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.9 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 0.6 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.2 | 0.6 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.5 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.4 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.4 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.5 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0072097 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.8 | GO:2000391 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.5 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.4 | GO:0070944 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.2 | GO:2000987 | cell communication by chemical coupling(GO:0010643) positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.3 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.5 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.2 | GO:2001280 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.5 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.2 | GO:1903314 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0060345 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.3 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.4 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 0.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.7 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.8 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:1903598 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 2.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.7 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 5.7 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.5 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.0 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.5 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 1.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 1.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.6 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.3 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 2.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 6.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |