Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
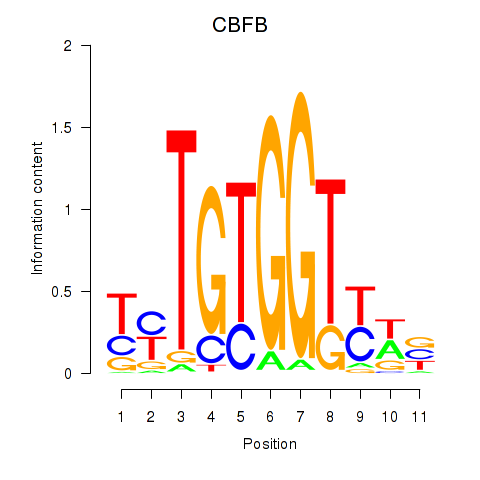
Results for CBFB
Z-value: 0.48
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67063262_67063364 | 0.89 | 1.8e-02 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209859510 | 0.55 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_+_173955327 | 0.40 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr1_+_196621002 | 0.37 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr11_+_114310237 | 0.33 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr7_+_7196565 | 0.33 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr1_+_196621156 | 0.31 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr21_+_35107346 | 0.31 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr4_-_156297949 | 0.30 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr15_+_49715449 | 0.28 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr5_+_131409476 | 0.28 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr14_-_35183886 | 0.26 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr1_-_240775447 | 0.25 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr5_-_39203093 | 0.22 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr1_+_17634689 | 0.20 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr9_-_130541017 | 0.20 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr5_+_35856951 | 0.19 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr15_-_64665911 | 0.19 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr8_-_96281419 | 0.17 |
ENST00000286688.5
|
C8orf37
|
chromosome 8 open reading frame 37 |
| chr8_+_104384616 | 0.17 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr5_-_137878887 | 0.16 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr6_-_113953705 | 0.16 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr2_-_127977654 | 0.16 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr2_-_87248975 | 0.15 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr1_+_89246647 | 0.14 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr6_+_30908747 | 0.14 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr17_-_39183452 | 0.14 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr15_+_49715293 | 0.12 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr6_+_27925019 | 0.12 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr11_-_58343319 | 0.11 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr11_+_114310164 | 0.11 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr12_-_53575120 | 0.11 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr13_-_103019744 | 0.11 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr13_+_76210448 | 0.10 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr22_+_42665742 | 0.10 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr19_-_14049184 | 0.10 |
ENST00000339560.5
|
PODNL1
|
podocan-like 1 |
| chr5_-_108745689 | 0.08 |
ENST00000361189.2
|
PJA2
|
praja ring finger 2, E3 ubiquitin protein ligase |
| chr1_-_184723942 | 0.07 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr6_-_75912508 | 0.07 |
ENST00000416123.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr12_-_121972556 | 0.07 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chrX_+_115567767 | 0.07 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr12_-_4754318 | 0.07 |
ENST00000536414.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr17_+_32582293 | 0.07 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr12_-_54778244 | 0.06 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr11_+_114310102 | 0.06 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr3_+_157154578 | 0.06 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_1912084 | 0.06 |
ENST00000391480.1
|
C11orf89
|
chromosome 11 open reading frame 89 |
| chr5_+_67576109 | 0.06 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_4754356 | 0.06 |
ENST00000540967.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr1_+_66796401 | 0.06 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_+_53088885 | 0.06 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr21_-_36421626 | 0.06 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr6_-_34524049 | 0.05 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr11_-_18343669 | 0.05 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr11_-_66103867 | 0.05 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_+_207024306 | 0.04 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr8_+_67624653 | 0.04 |
ENST00000521198.2
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr18_-_53089538 | 0.04 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr7_+_120628731 | 0.04 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr18_-_53089723 | 0.04 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr12_-_14849470 | 0.04 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr12_-_54778444 | 0.04 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr13_-_28674693 | 0.04 |
ENST00000537084.1
ENST00000241453.7 ENST00000380982.4 |
FLT3
|
fms-related tyrosine kinase 3 |
| chr2_-_158300556 | 0.04 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr15_-_64386120 | 0.04 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr14_-_85996332 | 0.04 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr14_-_59951112 | 0.04 |
ENST00000247194.4
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr20_+_54987305 | 0.03 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr15_-_64385981 | 0.03 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr3_+_156799587 | 0.03 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr14_-_59950724 | 0.02 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr1_-_184006829 | 0.02 |
ENST00000361927.4
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr21_-_36421401 | 0.02 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr11_+_18343800 | 0.02 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr8_-_60031762 | 0.01 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr6_+_45296391 | 0.01 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr12_-_54778471 | 0.01 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr20_+_54987168 | 0.01 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr7_+_117119187 | 0.01 |
ENST00000446805.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr17_-_34417479 | 0.00 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |