Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
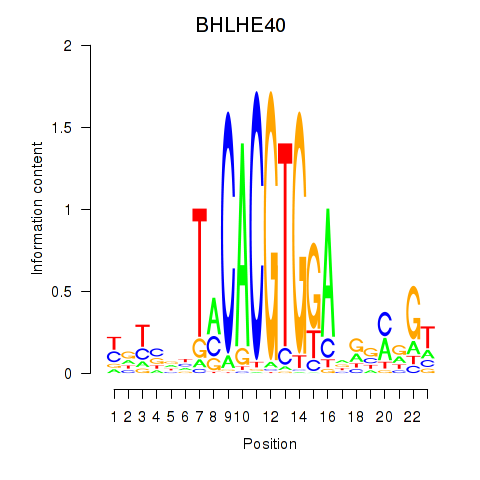
Results for BHLHE40
Z-value: 0.59
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | -0.87 | 2.6e-02 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_190649107 | 0.62 |
ENST00000441310.2
ENST00000409985.1 ENST00000446877.1 ENST00000418224.3 ENST00000409823.3 ENST00000374826.4 ENST00000424766.1 ENST00000447232.2 ENST00000432292.3 |
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr2_-_190649082 | 0.44 |
ENST00000392350.3
ENST00000392349.4 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr1_+_203830703 | 0.43 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr5_-_145214893 | 0.41 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr1_-_86174065 | 0.38 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr17_+_53046096 | 0.37 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr2_-_136743039 | 0.35 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr5_+_94982435 | 0.35 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr4_-_57301748 | 0.34 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr2_-_190649023 | 0.34 |
ENST00000409519.1
ENST00000458355.1 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr21_+_27107672 | 0.31 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr19_-_37064145 | 0.31 |
ENST00000591340.1
ENST00000334116.7 |
ZNF529
|
zinc finger protein 529 |
| chr15_+_77713222 | 0.30 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr8_-_99129338 | 0.29 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr22_+_48885379 | 0.29 |
ENST00000336769.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr14_-_45722605 | 0.27 |
ENST00000310806.4
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr7_-_960521 | 0.26 |
ENST00000437486.1
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr21_-_27107344 | 0.26 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr8_-_99129384 | 0.26 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr12_+_93861282 | 0.25 |
ENST00000552217.1
ENST00000393128.4 ENST00000547098.1 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr5_-_36152031 | 0.25 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr8_+_26240414 | 0.25 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr8_+_109455830 | 0.24 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr2_+_207630081 | 0.24 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr14_-_45722360 | 0.24 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr12_+_66217911 | 0.24 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr2_-_136743169 | 0.24 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr21_-_27107283 | 0.24 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr4_-_129207942 | 0.23 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr12_+_93861264 | 0.23 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chrX_+_23685563 | 0.23 |
ENST00000379341.4
|
PRDX4
|
peroxiredoxin 4 |
| chr2_-_179343226 | 0.22 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr8_+_109455845 | 0.22 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr4_-_129208030 | 0.22 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr8_+_125985531 | 0.22 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr2_+_201676908 | 0.21 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr5_+_179078298 | 0.21 |
ENST00000418535.2
ENST00000425471.1 |
AC136604.1
|
Uncharacterized protein |
| chr19_-_695427 | 0.20 |
ENST00000329267.7
|
PRSS57
|
protease, serine, 57 |
| chr19_+_51774540 | 0.20 |
ENST00000600813.1
|
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr7_-_229557 | 0.19 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr1_+_207070775 | 0.19 |
ENST00000391929.3
ENST00000294984.2 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr2_-_47142884 | 0.19 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr10_+_49514698 | 0.18 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr9_+_37120536 | 0.18 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chrX_+_23685653 | 0.18 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr2_-_136743436 | 0.18 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr10_-_35379241 | 0.17 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr10_-_35379524 | 0.17 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr4_+_57301896 | 0.17 |
ENST00000514888.1
ENST00000264221.2 ENST00000505164.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr11_+_34664014 | 0.16 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr6_-_109804412 | 0.16 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr6_-_36842784 | 0.15 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr2_+_183989157 | 0.15 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr7_+_141463897 | 0.15 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr4_+_57302297 | 0.14 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr4_+_119200215 | 0.14 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr17_-_61850894 | 0.14 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr21_-_30391636 | 0.14 |
ENST00000493196.1
|
RWDD2B
|
RWD domain containing 2B |
| chr4_+_41983713 | 0.14 |
ENST00000333141.5
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1 |
| chr2_-_47143160 | 0.13 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr6_-_5261141 | 0.13 |
ENST00000330636.4
ENST00000500576.2 |
LYRM4
|
LYR motif containing 4 |
| chr1_+_231376941 | 0.13 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr9_-_99540328 | 0.13 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr19_+_51774520 | 0.13 |
ENST00000595566.1
ENST00000594261.1 |
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr19_+_41497178 | 0.13 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr4_-_159094194 | 0.13 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr6_+_31865552 | 0.13 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr21_-_27107881 | 0.12 |
ENST00000400090.3
ENST00000400087.3 ENST00000400093.3 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr5_-_114880533 | 0.12 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr21_-_45079341 | 0.12 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr5_+_89770696 | 0.12 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr6_+_144164455 | 0.12 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr17_-_53046058 | 0.12 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr1_-_113498943 | 0.12 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr6_-_151773232 | 0.12 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr19_+_7660716 | 0.11 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr19_+_12305824 | 0.11 |
ENST00000415793.1
ENST00000440004.1 ENST00000426044.1 |
CTD-2666L21.1
|
CTD-2666L21.1 |
| chr17_+_48796905 | 0.11 |
ENST00000505658.1
ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_-_200589859 | 0.11 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr12_+_65004292 | 0.11 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr5_+_89770664 | 0.11 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr4_-_100009856 | 0.11 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chrX_-_16887963 | 0.10 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr13_+_35516390 | 0.10 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr2_-_179343268 | 0.10 |
ENST00000424785.2
|
FKBP7
|
FK506 binding protein 7 |
| chr17_-_7193711 | 0.10 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr9_-_100881466 | 0.10 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr10_-_21435488 | 0.10 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr12_-_25102252 | 0.10 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr16_-_79804435 | 0.09 |
ENST00000562921.1
ENST00000566729.1 |
RP11-345M22.2
|
RP11-345M22.2 |
| chr17_+_72199721 | 0.09 |
ENST00000439590.2
ENST00000311111.6 ENST00000584577.1 ENST00000534490.1 ENST00000528433.2 ENST00000533498.1 |
RPL38
|
ribosomal protein L38 |
| chr8_+_55047763 | 0.09 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr1_+_44514040 | 0.09 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr17_-_10600818 | 0.09 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chr2_+_183989083 | 0.09 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr11_-_61560053 | 0.09 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr9_-_95640218 | 0.08 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr3_+_133293278 | 0.08 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr13_+_111972980 | 0.08 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr1_-_26700943 | 0.08 |
ENST00000416125.1
|
ZNF683
|
zinc finger protein 683 |
| chr4_+_188916918 | 0.08 |
ENST00000509524.1
ENST00000326866.4 |
ZFP42
|
ZFP42 zinc finger protein |
| chr10_+_101491968 | 0.08 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr7_-_151574191 | 0.08 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_-_6019455 | 0.07 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr7_-_100493744 | 0.07 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr1_+_201708992 | 0.07 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chrX_+_129040122 | 0.07 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr2_-_112642267 | 0.07 |
ENST00000341068.3
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr3_+_111578640 | 0.07 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr5_-_89770171 | 0.06 |
ENST00000514906.1
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr16_-_56701933 | 0.06 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr13_+_100741269 | 0.06 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr12_-_58165870 | 0.06 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr6_-_5260963 | 0.06 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chrX_+_55101495 | 0.06 |
ENST00000374974.3
ENST00000374971.1 |
PAGE2B
|
P antigen family, member 2B |
| chr16_-_79804394 | 0.05 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr2_+_105654441 | 0.05 |
ENST00000258455.3
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr17_+_74733744 | 0.05 |
ENST00000586689.1
ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr15_-_35088340 | 0.05 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1 |
| chr1_+_210001309 | 0.04 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr10_-_49459800 | 0.04 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr14_+_73393040 | 0.04 |
ENST00000358377.2
ENST00000353777.3 ENST00000394234.2 ENST00000509153.1 ENST00000555042.1 |
DCAF4
|
DDB1 and CUL4 associated factor 4 |
| chr22_+_48885272 | 0.04 |
ENST00000402357.1
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr3_+_63638372 | 0.04 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr7_-_155326532 | 0.04 |
ENST00000406197.1
ENST00000321736.5 |
CNPY1
|
canopy FGF signaling regulator 1 |
| chr1_-_231376867 | 0.04 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr12_-_54673871 | 0.04 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr14_+_101359265 | 0.04 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr21_+_33784670 | 0.03 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr1_+_173793777 | 0.03 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr12_-_25101920 | 0.03 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr5_-_89770582 | 0.03 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr1_+_76262552 | 0.03 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4 |
| chr5_+_36152163 | 0.03 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr19_-_52133588 | 0.03 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chrX_+_66764375 | 0.03 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr3_+_108015370 | 0.03 |
ENST00000467282.1
|
HHLA2
|
HERV-H LTR-associating 2 |
| chr12_-_55042140 | 0.02 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr5_-_145214848 | 0.02 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr12_-_122710556 | 0.02 |
ENST00000446652.1
ENST00000541273.1 ENST00000353548.6 ENST00000267169.6 ENST00000464942.2 |
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr13_-_98829501 | 0.02 |
ENST00000267291.6
|
RNF113B
|
ring finger protein 113B |
| chr3_+_99536663 | 0.02 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr9_+_131684027 | 0.02 |
ENST00000426694.1
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr12_+_54674482 | 0.02 |
ENST00000547708.1
ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr20_+_31755934 | 0.02 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr17_-_8661860 | 0.02 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr3_-_157217328 | 0.02 |
ENST00000392832.2
ENST00000543418.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_-_47518498 | 0.02 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr17_-_13505219 | 0.02 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr15_-_23692381 | 0.01 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr10_-_6019552 | 0.01 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr8_+_58890917 | 0.01 |
ENST00000522992.1
|
RP11-1112C15.1
|
RP11-1112C15.1 |
| chr19_-_3786253 | 0.01 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chrX_+_129040094 | 0.01 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr8_-_48872686 | 0.01 |
ENST00000314191.2
ENST00000338368.3 |
PRKDC
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr2_-_10588630 | 0.01 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr6_+_44215603 | 0.01 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr22_+_22681656 | 0.00 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr1_+_118472343 | 0.00 |
ENST00000369441.3
ENST00000349139.5 |
WDR3
|
WD repeat domain 3 |
| chr6_-_154677866 | 0.00 |
ENST00000367220.4
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.2 | 0.8 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.2 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |