Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
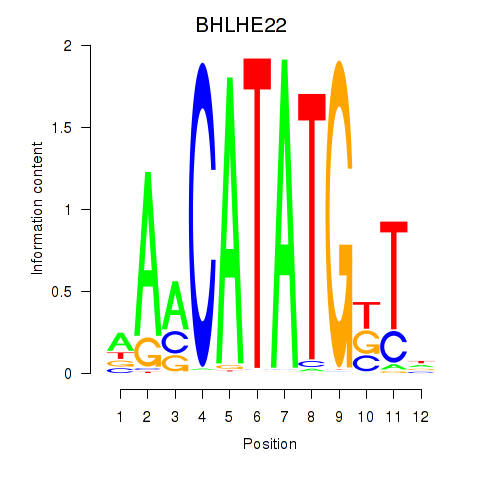
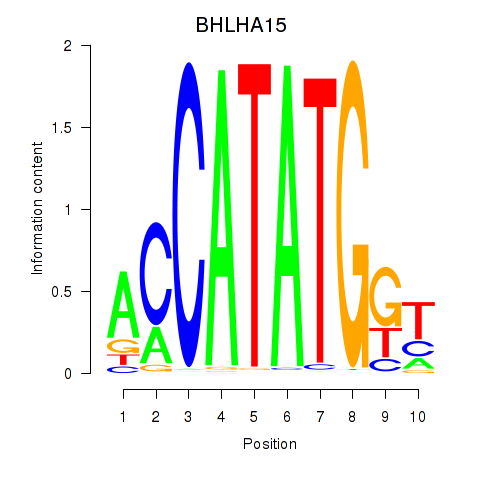
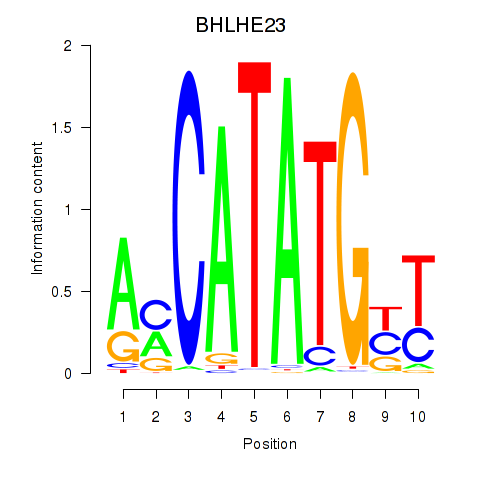
Results for BHLHE22_BHLHA15_BHLHE23
Z-value: 0.76
Transcription factors associated with BHLHE22_BHLHA15_BHLHE23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE22
|
ENSG00000180828.1 | basic helix-loop-helix family member e22 |
|
BHLHA15
|
ENSG00000180535.3 | basic helix-loop-helix family member a15 |
|
BHLHE23
|
ENSG00000125533.4 | basic helix-loop-helix family member e23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE22 | hg19_v2_chr8_+_65492756_65492814 | 0.73 | 9.9e-02 | Click! |
| BHLHA15 | hg19_v2_chr7_+_97840739_97840786 | -0.19 | 7.2e-01 | Click! |
Activity profile of BHLHE22_BHLHA15_BHLHE23 motif
Sorted Z-values of BHLHE22_BHLHA15_BHLHE23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_66918558 | 0.82 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr6_+_135502501 | 0.62 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_-_178984759 | 0.45 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr16_-_69385681 | 0.41 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr6_+_163148161 | 0.39 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr8_+_67782984 | 0.35 |
ENST00000396592.3
ENST00000422365.2 ENST00000492775.1 |
MCMDC2
|
minichromosome maintenance domain containing 2 |
| chr1_+_185126598 | 0.30 |
ENST00000450350.1
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr5_-_159766528 | 0.28 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chrX_+_70798261 | 0.28 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr7_-_101212244 | 0.26 |
ENST00000451953.1
ENST00000434537.1 ENST00000437900.1 |
LINC01007
|
long intergenic non-protein coding RNA 1007 |
| chr17_+_75316336 | 0.26 |
ENST00000591934.1
|
SEPT9
|
septin 9 |
| chr11_-_26743546 | 0.25 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr15_+_40650408 | 0.25 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr1_-_186344802 | 0.23 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr1_-_98515395 | 0.23 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr11_+_65405556 | 0.23 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr19_+_57874835 | 0.22 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr10_+_89124746 | 0.21 |
ENST00000465545.1
|
NUTM2D
|
NUT family member 2D |
| chr17_+_7211656 | 0.21 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_94673956 | 0.20 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr3_-_121379739 | 0.19 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr4_+_55095428 | 0.18 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_+_75713481 | 0.18 |
ENST00000308062.3
ENST00000464571.1 |
FRG2C
|
FSHD region gene 2 family, member C |
| chr14_-_107283278 | 0.17 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr1_+_81106951 | 0.16 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr3_-_114477962 | 0.15 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr15_+_65843130 | 0.15 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr13_+_28813645 | 0.15 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr8_+_38239882 | 0.15 |
ENST00000607047.1
|
RP11-350N15.5
|
RP11-350N15.5 |
| chr1_+_244214577 | 0.13 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr7_-_38403077 | 0.12 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr7_-_27205136 | 0.12 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr20_+_33292507 | 0.12 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr3_-_114477787 | 0.11 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_-_101724989 | 0.11 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chrX_+_105855160 | 0.11 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr17_+_46048497 | 0.11 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr19_+_42041702 | 0.11 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr21_-_36421401 | 0.10 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr14_+_32414059 | 0.10 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr1_-_168464875 | 0.10 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr17_-_73874654 | 0.09 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr15_-_55700216 | 0.09 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr14_-_69444957 | 0.09 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr5_-_111312622 | 0.08 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr7_-_48068699 | 0.08 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr1_+_186344945 | 0.08 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr3_+_72201910 | 0.08 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr16_+_31119615 | 0.08 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr5_+_173316341 | 0.08 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr18_-_70305745 | 0.08 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chr11_-_115127611 | 0.08 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr8_-_135522425 | 0.07 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chrX_+_107288239 | 0.07 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_+_7211280 | 0.07 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr1_-_205091115 | 0.07 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chrX_+_107288197 | 0.07 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_-_108278456 | 0.07 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr11_+_66008698 | 0.07 |
ENST00000531597.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr4_+_74301880 | 0.06 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr10_-_15902449 | 0.06 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr3_-_55515400 | 0.06 |
ENST00000497027.1
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr15_+_85144217 | 0.06 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr19_+_42041860 | 0.06 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr3_+_62304648 | 0.05 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr2_+_219840955 | 0.05 |
ENST00000598002.1
ENST00000432733.1 |
LINC00608
|
long intergenic non-protein coding RNA 608 |
| chr3_-_55515202 | 0.05 |
ENST00000482079.1
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr3_-_150264272 | 0.05 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chrX_-_15402498 | 0.05 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr9_-_10612703 | 0.04 |
ENST00000463477.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr16_+_72142195 | 0.04 |
ENST00000563819.1
ENST00000567142.2 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr10_+_17794251 | 0.04 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr17_-_47286729 | 0.04 |
ENST00000300406.2
ENST00000511277.1 ENST00000511673.1 |
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr1_+_180897269 | 0.04 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr15_+_63354769 | 0.04 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr6_+_4087664 | 0.04 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr7_-_48068671 | 0.04 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr13_+_49551020 | 0.04 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr4_-_113207048 | 0.04 |
ENST00000361717.3
|
TIFA
|
TRAF-interacting protein with forkhead-associated domain |
| chr1_+_12524965 | 0.04 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_-_44818599 | 0.03 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr4_-_141075330 | 0.03 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr12_+_2912363 | 0.03 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr3_-_52719546 | 0.03 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr19_-_52227221 | 0.03 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr7_-_97881429 | 0.03 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr3_+_185300270 | 0.03 |
ENST00000430355.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr11_+_120107344 | 0.03 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr6_+_116442320 | 0.03 |
ENST00000430695.1
|
AL121963.1
|
Uncharacterized protein |
| chr5_+_71475449 | 0.03 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr16_-_4896205 | 0.03 |
ENST00000589389.1
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr12_+_79371565 | 0.03 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr12_+_58166431 | 0.03 |
ENST00000333012.5
|
METTL21B
|
methyltransferase like 21B |
| chr1_+_100810575 | 0.02 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chrX_-_19817869 | 0.02 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr10_+_99627889 | 0.02 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr3_-_33700589 | 0.02 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_-_56492989 | 0.02 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr8_+_21823726 | 0.02 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr11_-_44972418 | 0.02 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr2_+_210444298 | 0.02 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_+_80858418 | 0.02 |
ENST00000574422.1
|
TBCD
|
tubulin folding cofactor D |
| chr15_-_55700522 | 0.02 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr1_+_170633047 | 0.02 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr7_+_116502605 | 0.02 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr8_-_67976509 | 0.02 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr12_-_95397442 | 0.02 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr4_+_115519894 | 0.02 |
ENST00000507710.1
|
UGT8
|
UDP glycosyltransferase 8 |
| chr10_-_74114714 | 0.02 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr15_-_55700457 | 0.01 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr9_+_12775011 | 0.01 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr5_-_171433579 | 0.01 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chrX_+_107334895 | 0.01 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr9_+_108424738 | 0.01 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr2_+_162480918 | 0.01 |
ENST00000272716.5
ENST00000446997.1 |
SLC4A10
|
solute carrier family 4, sodium bicarbonate transporter, member 10 |
| chr11_+_94501497 | 0.01 |
ENST00000317829.8
ENST00000317837.9 ENST00000433060.2 |
AMOTL1
|
angiomotin like 1 |
| chr16_+_14726672 | 0.01 |
ENST00000261658.2
ENST00000563971.1 |
BFAR
|
bifunctional apoptosis regulator |
| chr14_-_51297837 | 0.01 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr4_+_55095264 | 0.01 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr14_-_51297197 | 0.01 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr4_-_100575781 | 0.01 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr2_+_181845843 | 0.01 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr11_-_44972476 | 0.01 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr9_+_125796806 | 0.01 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr6_-_112115103 | 0.01 |
ENST00000462598.3
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr2_-_32490859 | 0.01 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chrX_+_128913906 | 0.01 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chrX_-_84363974 | 0.01 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr15_-_98417780 | 0.01 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr4_-_159094194 | 0.01 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr2_-_227050079 | 0.00 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chrX_-_13835147 | 0.00 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr2_-_32490801 | 0.00 |
ENST00000360906.5
ENST00000342905.6 |
NLRC4
|
NLR family, CARD domain containing 4 |
| chr13_+_111766897 | 0.00 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr12_-_91573132 | 0.00 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr17_-_47286244 | 0.00 |
ENST00000503070.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr12_+_50794891 | 0.00 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr10_+_18629628 | 0.00 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chrX_+_69509927 | 0.00 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr5_-_171433819 | 0.00 |
ENST00000296933.6
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chrX_-_13835461 | 0.00 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr11_-_62359027 | 0.00 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr5_-_114631958 | 0.00 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr2_+_108994466 | 0.00 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE22_BHLHA15_BHLHE23
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.2 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0061348 | chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) melanocyte proliferation(GO:0097325) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |