Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for BARHL2
Z-value: 0.88
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.7 | BarH like homeobox 2 |
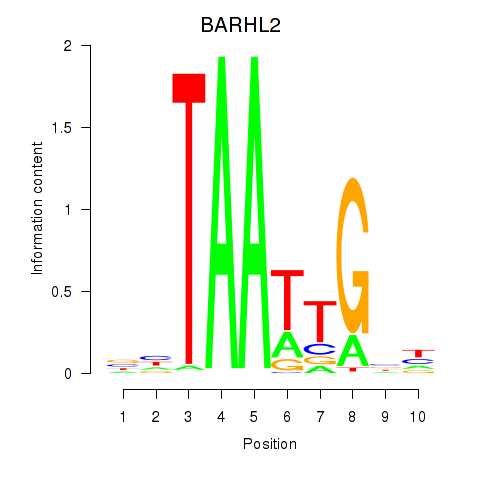
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_53412368 | 0.86 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr8_+_94752349 | 0.77 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr8_-_150563 | 0.56 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr15_+_58702742 | 0.52 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr17_+_61151306 | 0.50 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr13_+_110958124 | 0.48 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr1_+_159931002 | 0.48 |
ENST00000443364.1
ENST00000423943.1 |
RP11-48O20.4
|
long intergenic non-protein coding RNA 1133 |
| chr6_-_26032288 | 0.43 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr21_-_19858196 | 0.39 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr8_+_19536083 | 0.39 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr17_-_39341594 | 0.38 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr12_-_91546926 | 0.37 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_+_76037081 | 0.36 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr1_+_59486129 | 0.35 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr14_-_92247032 | 0.35 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr6_+_131571535 | 0.35 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr8_+_38831683 | 0.31 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr2_-_114647327 | 0.30 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr14_-_91294472 | 0.29 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chr2_-_87248975 | 0.29 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr3_-_71114066 | 0.29 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr4_-_89442940 | 0.28 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr7_-_84122033 | 0.27 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_-_168464875 | 0.27 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr12_-_76879852 | 0.26 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr13_-_99910673 | 0.26 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chrX_+_591524 | 0.26 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr7_-_81635106 | 0.26 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_-_91573132 | 0.26 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr4_-_105416039 | 0.25 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr17_-_29624343 | 0.25 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr5_-_145483932 | 0.25 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr3_+_174158732 | 0.24 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr14_+_73706308 | 0.24 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr2_-_203735484 | 0.24 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr4_+_70894130 | 0.24 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr20_+_31870927 | 0.24 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr4_-_142253761 | 0.23 |
ENST00000511213.1
|
RP11-362F19.1
|
RP11-362F19.1 |
| chr17_-_55162360 | 0.23 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr9_-_93405352 | 0.23 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr12_-_120241187 | 0.23 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_-_1208851 | 0.23 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr3_+_156799587 | 0.23 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chrX_-_100129128 | 0.22 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr4_+_71019903 | 0.22 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr16_+_22518495 | 0.22 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr12_-_75784669 | 0.21 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr10_-_33405600 | 0.21 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr5_+_31193847 | 0.21 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr11_+_12308447 | 0.21 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr2_-_203735586 | 0.20 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_+_116955659 | 0.20 |
ENST00000552992.1
|
RP11-148B3.1
|
RP11-148B3.1 |
| chr14_-_30766223 | 0.20 |
ENST00000549360.1
ENST00000508469.2 |
CTD-2251F13.1
|
CTD-2251F13.1 |
| chr9_+_130160440 | 0.20 |
ENST00000439597.1
ENST00000423934.1 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr2_+_196313239 | 0.20 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr22_-_31688381 | 0.20 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr16_+_21244986 | 0.20 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr7_+_116593536 | 0.20 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr17_-_46799872 | 0.19 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr1_+_186265399 | 0.19 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr2_+_87808725 | 0.19 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr8_-_109799793 | 0.19 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr10_+_24755416 | 0.19 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr5_+_140254884 | 0.19 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr19_+_39687596 | 0.19 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr6_+_13182751 | 0.19 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_-_7800067 | 0.18 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr12_+_25055243 | 0.18 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr5_-_146781153 | 0.18 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr9_-_95244781 | 0.18 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr7_-_14026123 | 0.17 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr10_-_63995871 | 0.17 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr10_+_135050908 | 0.17 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr8_-_124553437 | 0.17 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr4_-_109541610 | 0.17 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr16_-_21442874 | 0.17 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chrX_-_49056635 | 0.17 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chrM_+_9207 | 0.17 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr9_+_67977438 | 0.17 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr11_-_795286 | 0.17 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr5_-_74162739 | 0.16 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr11_-_795400 | 0.16 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr6_+_155538093 | 0.16 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr7_-_16921601 | 0.16 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr14_-_107283278 | 0.16 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr10_+_106034637 | 0.16 |
ENST00000401888.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr8_-_42396185 | 0.16 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_+_70916119 | 0.16 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr17_+_1666108 | 0.16 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr13_+_24144509 | 0.16 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_120498357 | 0.15 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr12_-_121972556 | 0.15 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chrX_+_22056165 | 0.15 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr5_-_87516448 | 0.15 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr4_+_22999152 | 0.15 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr7_+_150065278 | 0.15 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr6_-_56492816 | 0.15 |
ENST00000522360.1
|
DST
|
dystonin |
| chr3_-_178984759 | 0.15 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chrM_+_10053 | 0.15 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr12_+_26348246 | 0.15 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr18_+_29027696 | 0.15 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr10_+_24497704 | 0.15 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr2_-_73869508 | 0.15 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr2_-_203736150 | 0.15 |
ENST00000457524.1
ENST00000421334.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr11_+_77300669 | 0.14 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr12_+_107712173 | 0.14 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr2_+_158114051 | 0.14 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr12_-_86650077 | 0.14 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr9_-_69229650 | 0.14 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr16_+_14280564 | 0.14 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr9_-_79267432 | 0.14 |
ENST00000424866.1
|
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr4_+_144312659 | 0.14 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr13_-_30160925 | 0.13 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_+_172309634 | 0.13 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr6_-_26199499 | 0.13 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr12_+_52056548 | 0.13 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr6_-_97731019 | 0.13 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr14_+_61449197 | 0.13 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr9_-_34620440 | 0.13 |
ENST00000421919.1
ENST00000378911.3 ENST00000477738.2 ENST00000341694.2 ENST00000259632.7 ENST00000378913.2 ENST00000378916.4 ENST00000447983.2 |
DCTN3
|
dynactin 3 (p22) |
| chr11_+_92085262 | 0.13 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr7_-_14029283 | 0.13 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr21_+_38792602 | 0.13 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr20_+_49348109 | 0.13 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr12_+_72058130 | 0.13 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr1_-_15497774 | 0.13 |
ENST00000424792.1
ENST00000376005.3 |
C1orf195
|
chromosome 1 open reading frame 195 |
| chr1_+_40713573 | 0.13 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr16_+_72088376 | 0.12 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr6_-_139613269 | 0.12 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr8_-_116681123 | 0.12 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_-_64684672 | 0.12 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr2_+_133874577 | 0.12 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr4_-_48782259 | 0.12 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr19_+_58570605 | 0.12 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chr4_-_185139062 | 0.12 |
ENST00000296741.2
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr7_-_130080977 | 0.12 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr6_+_26087509 | 0.12 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr11_+_76092353 | 0.12 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr11_-_26743546 | 0.12 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr21_+_43619796 | 0.12 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr6_-_113754604 | 0.12 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr3_-_71777824 | 0.12 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr2_-_231989808 | 0.12 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chrX_+_100224676 | 0.12 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr14_-_21566731 | 0.12 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr11_+_6411636 | 0.12 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr14_+_23340822 | 0.12 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr7_-_14026063 | 0.12 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr16_-_2031464 | 0.11 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr7_-_112635675 | 0.11 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr20_-_45530365 | 0.11 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr5_+_140855495 | 0.11 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr8_+_107738240 | 0.11 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr2_-_55237484 | 0.11 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr8_+_67405794 | 0.11 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr12_-_89746264 | 0.11 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr17_-_38928414 | 0.11 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr4_-_122148620 | 0.11 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr18_-_48351743 | 0.11 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr13_-_99910620 | 0.11 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr12_+_20963647 | 0.10 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_-_111927062 | 0.10 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_-_14029515 | 0.10 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chrX_+_22050546 | 0.10 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr14_-_70883708 | 0.10 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr2_-_145277569 | 0.10 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_10435032 | 0.10 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr2_-_177502254 | 0.10 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr1_-_11865351 | 0.10 |
ENST00000413656.1
ENST00000376585.1 ENST00000423400.1 ENST00000431243.1 |
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr9_-_95298254 | 0.10 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr4_-_70653673 | 0.10 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr19_-_10445399 | 0.10 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_+_11696464 | 0.10 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr7_+_129984630 | 0.10 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr2_+_54342533 | 0.10 |
ENST00000406041.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr11_-_71823715 | 0.10 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_+_110011571 | 0.10 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr15_-_54267147 | 0.10 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr1_+_89829610 | 0.10 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr19_-_44259136 | 0.10 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr14_+_20187174 | 0.10 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr20_+_56964253 | 0.10 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr6_-_116833500 | 0.09 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chrX_+_49832231 | 0.09 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr9_-_125240235 | 0.09 |
ENST00000259357.2
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1 |
| chr18_+_20714525 | 0.09 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr1_+_2066252 | 0.09 |
ENST00000466352.1
|
PRKCZ
|
protein kinase C, zeta |
| chr9_-_95298314 | 0.09 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr5_+_140593509 | 0.09 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr4_-_48136217 | 0.09 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr4_-_143226979 | 0.09 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chrX_+_38660682 | 0.09 |
ENST00000457894.1
|
MID1IP1
|
MID1 interacting protein 1 |
| chr8_+_125486939 | 0.09 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr8_-_116681221 | 0.09 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_-_225266802 | 0.09 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124B |
| chr12_+_14572070 | 0.09 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr3_-_47934234 | 0.09 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr10_-_49860525 | 0.09 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr12_-_102591604 | 0.09 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr5_-_86534822 | 0.09 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr12_-_27235447 | 0.09 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.0 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) |
| 0.0 | 0.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.2 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.2 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.2 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0015254 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |