Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for BACH2
Z-value: 0.52
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BTB domain and CNC homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH2 | hg19_v2_chr6_-_91006627_91006641 | 0.77 | 7.5e-02 | Click! |
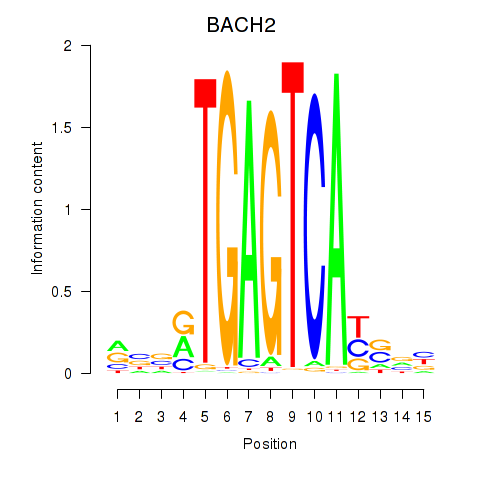
Activity profile of BACH2 motif
Sorted Z-values of BACH2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39264072 | 0.69 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr1_+_154378049 | 0.66 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr9_-_34662651 | 0.64 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr19_-_6057282 | 0.43 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr14_+_77425972 | 0.43 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr22_+_22676808 | 0.40 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr14_+_38065052 | 0.39 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr3_+_11267691 | 0.38 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr17_+_80317121 | 0.37 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr15_+_41062159 | 0.35 |
ENST00000344320.6
|
C15orf62
|
chromosome 15 open reading frame 62 |
| chr1_-_12679171 | 0.35 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr7_-_130597935 | 0.33 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr6_-_30685214 | 0.33 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr1_+_156096336 | 0.32 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr3_-_46608010 | 0.31 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr9_-_91793675 | 0.30 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr19_+_7953417 | 0.30 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr11_-_119066545 | 0.30 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr19_+_10959043 | 0.29 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr20_+_48909240 | 0.28 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr11_+_46316677 | 0.28 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr1_-_219615984 | 0.28 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr4_+_108746282 | 0.28 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr8_+_22844995 | 0.28 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr14_-_51863853 | 0.27 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr16_+_30077098 | 0.26 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_-_223853425 | 0.26 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr4_+_159727222 | 0.25 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr10_+_127661942 | 0.24 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr4_+_159727272 | 0.24 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr3_+_187930429 | 0.24 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_62457371 | 0.23 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr1_+_36621174 | 0.22 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_-_39538550 | 0.22 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr2_+_220144168 | 0.22 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_-_6662919 | 0.21 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr17_+_37030127 | 0.21 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr16_-_1429674 | 0.20 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr19_-_44008863 | 0.20 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr11_-_119993979 | 0.20 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr5_+_175288631 | 0.20 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr6_+_41604747 | 0.20 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr5_-_175843524 | 0.19 |
ENST00000502877.1
|
CLTB
|
clathrin, light chain B |
| chr17_-_39769005 | 0.18 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr11_-_62323702 | 0.18 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr12_+_10365082 | 0.18 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_6341724 | 0.18 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr14_+_105147464 | 0.18 |
ENST00000540171.2
|
RP11-982M15.6
|
RP11-982M15.6 |
| chr5_-_176923803 | 0.17 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr1_-_223853348 | 0.16 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr1_+_156095951 | 0.16 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr1_+_156084461 | 0.16 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr1_+_223889310 | 0.16 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr16_+_30077055 | 0.16 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_+_184016986 | 0.16 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr6_+_2988933 | 0.16 |
ENST00000597787.1
|
LINC01011
|
long intergenic non-protein coding RNA 1011 |
| chrX_-_48901012 | 0.16 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr5_-_175843569 | 0.16 |
ENST00000310418.4
ENST00000345807.2 |
CLTB
|
clathrin, light chain B |
| chr8_+_22844913 | 0.15 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chrX_-_48937531 | 0.15 |
ENST00000473974.1
ENST00000475880.1 ENST00000396681.4 ENST00000553851.1 ENST00000471338.1 ENST00000476728.1 ENST00000376368.2 ENST00000485908.1 ENST00000376372.3 ENST00000376358.3 |
WDR45
AF196779.12
|
WD repeat domain 45 WD repeat domain phosphoinositide-interacting protein 4 |
| chr11_+_35639735 | 0.15 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr16_-_1429627 | 0.15 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr1_+_154966058 | 0.15 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr17_-_18161870 | 0.15 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr11_-_2924720 | 0.15 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chrX_+_37765364 | 0.14 |
ENST00000567273.1
|
AL121578.2
|
AL121578.2 |
| chr7_+_134528635 | 0.14 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr16_+_28996416 | 0.14 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr6_+_56819773 | 0.14 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr10_+_121410882 | 0.14 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr11_-_8739383 | 0.14 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr10_+_17270214 | 0.14 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr1_+_19638788 | 0.14 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr12_-_95009837 | 0.13 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr16_+_57662596 | 0.13 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_56595196 | 0.13 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr1_-_158656488 | 0.13 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr12_+_52695617 | 0.13 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr3_+_57094469 | 0.13 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr12_-_56101647 | 0.13 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chr1_-_179112189 | 0.13 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr11_-_66104237 | 0.12 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_-_9781068 | 0.12 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr3_-_122512619 | 0.12 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr14_+_69726656 | 0.12 |
ENST00000337827.4
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr12_-_53297432 | 0.12 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr5_+_141016969 | 0.12 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chrX_-_48937684 | 0.12 |
ENST00000465382.1
ENST00000423215.2 |
WDR45
|
WD repeat domain 45 |
| chrX_-_48937503 | 0.12 |
ENST00000322995.8
|
WDR45
|
WD repeat domain 45 |
| chr12_+_56732658 | 0.12 |
ENST00000228534.4
|
IL23A
|
interleukin 23, alpha subunit p19 |
| chr2_+_220143989 | 0.12 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_-_156675368 | 0.12 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_+_25799008 | 0.12 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr1_+_36621697 | 0.12 |
ENST00000373150.4
ENST00000373151.2 |
MAP7D1
|
MAP7 domain containing 1 |
| chr11_+_10477733 | 0.11 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr16_+_30968615 | 0.11 |
ENST00000262519.8
|
SETD1A
|
SET domain containing 1A |
| chr5_+_150020240 | 0.11 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr1_+_36554470 | 0.11 |
ENST00000373178.4
|
ADPRHL2
|
ADP-ribosylhydrolase like 2 |
| chr5_+_179247759 | 0.11 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr1_-_179112173 | 0.11 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr18_+_21452804 | 0.11 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr1_+_36621529 | 0.11 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_+_75315654 | 0.11 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr17_+_75315534 | 0.11 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr16_+_67571351 | 0.11 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr15_-_72521017 | 0.11 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr1_+_154377669 | 0.11 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr19_+_38794797 | 0.11 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr2_-_145275109 | 0.11 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_67802549 | 0.11 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr12_+_10365404 | 0.10 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr3_+_183903811 | 0.10 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr5_+_150020214 | 0.10 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr12_-_118490403 | 0.10 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr9_+_140135665 | 0.10 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr19_-_10450287 | 0.10 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr5_+_149877334 | 0.10 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr18_-_74207146 | 0.10 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr1_+_155107820 | 0.10 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr13_-_41768654 | 0.10 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr6_-_24877490 | 0.10 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr17_-_27467418 | 0.10 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr11_-_102826434 | 0.10 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr17_+_79650962 | 0.09 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr5_-_138534071 | 0.09 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr2_+_201980827 | 0.09 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_13842559 | 0.09 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr5_-_141061777 | 0.09 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr20_+_48429233 | 0.09 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr7_+_150065278 | 0.09 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr6_-_31704282 | 0.09 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr16_+_22308717 | 0.09 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr7_+_100026406 | 0.09 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr16_+_58533951 | 0.09 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr4_-_987217 | 0.09 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr19_+_39279838 | 0.09 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr20_+_48429356 | 0.09 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_-_27503770 | 0.09 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr22_+_20861858 | 0.09 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr5_-_137090028 | 0.08 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr16_+_89988259 | 0.08 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr15_+_59903975 | 0.08 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr20_-_35807741 | 0.08 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr17_+_21191341 | 0.08 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr10_-_71892555 | 0.08 |
ENST00000307864.1
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr1_+_26606608 | 0.08 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr2_+_220144052 | 0.08 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_+_233562015 | 0.08 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chrX_-_129244655 | 0.07 |
ENST00000335997.7
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr20_-_634000 | 0.07 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr6_+_2988847 | 0.07 |
ENST00000380472.3
ENST00000605901.1 ENST00000454015.1 |
NQO2
LINC01011
|
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr3_+_48507621 | 0.07 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr12_-_8218997 | 0.07 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr11_-_65667997 | 0.07 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr9_-_139922726 | 0.07 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr16_-_75301886 | 0.07 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr16_-_122619 | 0.07 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr17_-_59668550 | 0.07 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr11_-_66103932 | 0.07 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr10_+_104155450 | 0.07 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_-_220117867 | 0.07 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr10_-_73611046 | 0.07 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr19_+_36630855 | 0.07 |
ENST00000589146.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr18_+_268148 | 0.07 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr12_+_7341759 | 0.07 |
ENST00000455147.2
ENST00000540398.1 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr11_-_65667884 | 0.07 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr1_-_153935791 | 0.07 |
ENST00000429040.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr2_-_65593784 | 0.07 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr10_+_88428206 | 0.07 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr12_-_53343560 | 0.07 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr9_-_130341268 | 0.07 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr3_-_48632593 | 0.07 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr3_+_112930387 | 0.07 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr17_+_79651007 | 0.07 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr9_-_114246635 | 0.07 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr3_-_49823941 | 0.07 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr19_+_36630454 | 0.06 |
ENST00000246533.3
|
CAPNS1
|
calpain, small subunit 1 |
| chr7_-_100026280 | 0.06 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr3_-_194090460 | 0.06 |
ENST00000428839.1
ENST00000347624.3 |
LRRC15
|
leucine rich repeat containing 15 |
| chr18_+_21452964 | 0.06 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr6_-_159239257 | 0.06 |
ENST00000337147.7
ENST00000392177.4 |
EZR
|
ezrin |
| chr6_+_56819895 | 0.06 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr22_+_20862321 | 0.06 |
ENST00000541476.1
ENST00000438962.1 |
MED15
|
mediator complex subunit 15 |
| chr1_+_26605618 | 0.06 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr3_+_69134080 | 0.06 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr17_+_55162453 | 0.06 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr9_-_127177703 | 0.06 |
ENST00000259457.3
ENST00000536392.1 ENST00000441097.1 |
PSMB7
|
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr16_+_30418910 | 0.06 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr7_+_143078379 | 0.06 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr3_+_38388251 | 0.06 |
ENST00000427323.1
ENST00000207870.3 ENST00000542835.1 |
XYLB
|
xylulokinase homolog (H. influenzae) |
| chr17_-_70417365 | 0.06 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr19_+_50706866 | 0.06 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr16_+_57662138 | 0.06 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr11_-_75201791 | 0.06 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr1_-_51796987 | 0.05 |
ENST00000262676.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr17_+_79071365 | 0.05 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chrX_-_129244454 | 0.05 |
ENST00000308167.5
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr12_-_53343633 | 0.05 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr11_-_65150103 | 0.05 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr5_-_176923846 | 0.05 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.6 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of activation of Janus kinase activity(GO:0010536) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 0.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |