Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
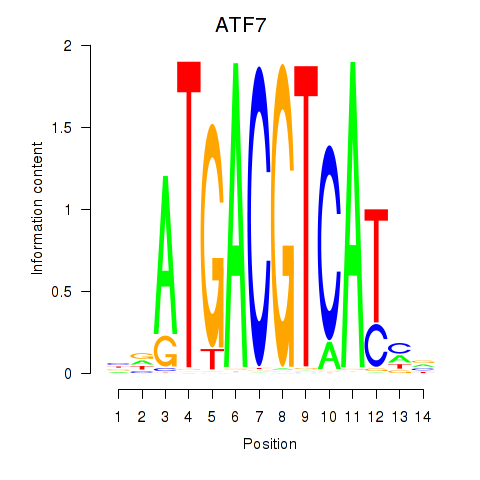
Results for ATF7
Z-value: 0.76
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | activating transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF7 | hg19_v2_chr12_-_54020107_54020199 | 0.54 | 2.6e-01 | Click! |
Activity profile of ATF7 motif
Sorted Z-values of ATF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_76829258 | 1.64 |
ENST00000588600.1
|
ATP9B
|
ATPase, class II, type 9B |
| chr18_+_76829441 | 0.75 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chrX_-_135056106 | 0.67 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr9_-_34665983 | 0.64 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr7_+_30174574 | 0.58 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chrX_-_135056216 | 0.53 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr7_-_124569864 | 0.51 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr12_+_100660940 | 0.45 |
ENST00000548392.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr4_-_104119528 | 0.42 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr9_-_99381660 | 0.40 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr3_+_62304712 | 0.40 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr3_+_169491171 | 0.38 |
ENST00000356716.4
|
MYNN
|
myoneurin |
| chr7_-_124569991 | 0.38 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr12_+_100660909 | 0.36 |
ENST00000549687.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr12_-_31882108 | 0.34 |
ENST00000281471.6
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr10_+_119000604 | 0.34 |
ENST00000298472.5
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine transporter), member 2 |
| chr6_-_106773291 | 0.33 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr3_+_62304648 | 0.32 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr7_+_30174668 | 0.31 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr7_+_30174426 | 0.31 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr14_+_61447927 | 0.30 |
ENST00000451406.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr10_-_105156198 | 0.30 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr21_-_30365136 | 0.30 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr6_-_27782548 | 0.30 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr3_+_158288942 | 0.29 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr3_+_169490834 | 0.29 |
ENST00000392733.1
|
MYNN
|
myoneurin |
| chr14_+_61447832 | 0.28 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr2_-_25194963 | 0.27 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr5_+_68513622 | 0.27 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr6_+_139456226 | 0.26 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr3_+_158288960 | 0.26 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr7_+_30960915 | 0.26 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr6_-_106773610 | 0.26 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr3_-_12587055 | 0.25 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr2_+_70142232 | 0.25 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr11_-_28129656 | 0.24 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr2_-_159313214 | 0.24 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr18_+_76829385 | 0.24 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr19_+_57874835 | 0.24 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr5_+_56469843 | 0.23 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr1_-_245026388 | 0.23 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr2_-_110962544 | 0.23 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr3_+_158288999 | 0.22 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr4_-_113558014 | 0.22 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chr4_-_113558079 | 0.22 |
ENST00000445203.2
|
C4orf21
|
chromosome 4 open reading frame 21 |
| chr4_-_104119488 | 0.22 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr11_+_102217936 | 0.22 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr3_+_170075436 | 0.21 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr4_+_85504075 | 0.20 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr1_+_168148169 | 0.20 |
ENST00000367833.2
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr1_+_153750622 | 0.20 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr11_+_17281900 | 0.19 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chrX_-_47863348 | 0.19 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr9_+_111696664 | 0.19 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr1_-_70671216 | 0.18 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr9_-_125027079 | 0.18 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr16_-_3068171 | 0.18 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr4_+_159131346 | 0.17 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr5_-_95297534 | 0.17 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr5_+_56469939 | 0.17 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr9_-_77643307 | 0.17 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chrX_+_155110956 | 0.17 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr3_+_140660634 | 0.17 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr10_-_18940501 | 0.16 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr4_+_113558272 | 0.16 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr12_-_93835665 | 0.16 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr11_-_82782952 | 0.16 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr11_+_3819049 | 0.15 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr8_-_82754193 | 0.15 |
ENST00000519817.1
ENST00000521773.1 ENST00000523757.1 |
SNX16
|
sorting nexin 16 |
| chr10_+_35484053 | 0.15 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr11_+_18344106 | 0.15 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr14_+_60716159 | 0.15 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr1_-_26700943 | 0.15 |
ENST00000416125.1
|
ZNF683
|
zinc finger protein 683 |
| chr4_-_146019287 | 0.15 |
ENST00000502847.1
ENST00000513054.1 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr12_+_100661156 | 0.15 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr11_-_33183006 | 0.15 |
ENST00000524827.1
ENST00000323959.4 ENST00000431742.2 |
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
| chrX_+_152224766 | 0.15 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr7_-_44924939 | 0.15 |
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B |
| chr3_+_140660743 | 0.15 |
ENST00000453248.2
|
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr17_+_33914424 | 0.15 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_-_154042205 | 0.14 |
ENST00000329463.5
|
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr8_+_94929168 | 0.14 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr5_+_56469775 | 0.14 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr12_-_93836028 | 0.14 |
ENST00000318066.2
|
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr2_-_112237835 | 0.14 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr11_+_82783097 | 0.14 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr2_+_220408724 | 0.14 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr1_-_115124257 | 0.14 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr8_+_26149007 | 0.14 |
ENST00000380737.3
ENST00000524169.1 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr8_+_30013813 | 0.14 |
ENST00000221114.3
|
DCTN6
|
dynactin 6 |
| chr17_+_29421900 | 0.14 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr7_+_28448995 | 0.14 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr4_+_78783674 | 0.14 |
ENST00000315567.8
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr8_+_30013875 | 0.13 |
ENST00000520829.1
|
DCTN6
|
dynactin 6 |
| chr1_+_2005126 | 0.13 |
ENST00000495347.1
|
PRKCZ
|
protein kinase C, zeta |
| chr12_-_123380610 | 0.13 |
ENST00000535765.1
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr13_+_49822041 | 0.13 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr7_+_100612904 | 0.13 |
ENST00000379442.3
ENST00000536621.1 |
MUC12
|
mucin 12, cell surface associated |
| chr3_-_195938256 | 0.13 |
ENST00000296326.3
|
ZDHHC19
|
zinc finger, DHHC-type containing 19 |
| chr15_-_72766533 | 0.13 |
ENST00000562573.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr6_-_106773491 | 0.13 |
ENST00000360666.4
|
ATG5
|
autophagy related 5 |
| chr14_+_96722539 | 0.13 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr7_+_23221438 | 0.12 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr6_+_30749649 | 0.12 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr14_+_88851874 | 0.12 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr4_+_146019421 | 0.12 |
ENST00000502586.1
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr14_+_60715928 | 0.12 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr3_+_51851612 | 0.12 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr8_+_94929077 | 0.12 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_-_67090825 | 0.12 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr5_+_70751442 | 0.12 |
ENST00000358731.4
ENST00000380675.2 |
BDP1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr2_+_37571845 | 0.12 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr18_+_2571510 | 0.12 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr6_-_27440460 | 0.12 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr1_+_29213678 | 0.12 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr15_+_42841008 | 0.11 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr16_+_11343475 | 0.11 |
ENST00000572173.1
|
RMI2
|
RecQ mediated genome instability 2 |
| chr5_+_112849373 | 0.11 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr14_-_101295407 | 0.11 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr11_-_31839488 | 0.11 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chr14_-_61447752 | 0.10 |
ENST00000555420.1
ENST00000553903.1 |
TRMT5
|
tRNA methyltransferase 5 |
| chr11_-_118122996 | 0.10 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr10_-_38265517 | 0.10 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr14_+_93389425 | 0.10 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr7_+_89975979 | 0.10 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr8_-_95274536 | 0.10 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr9_-_179018 | 0.10 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr20_+_17949684 | 0.10 |
ENST00000377709.1
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr4_+_667686 | 0.10 |
ENST00000505477.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr15_-_34394008 | 0.10 |
ENST00000527822.1
ENST00000528949.1 |
EMC7
|
ER membrane protein complex subunit 7 |
| chr15_-_72612470 | 0.10 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr5_-_178054105 | 0.10 |
ENST00000316308.4
|
CLK4
|
CDC-like kinase 4 |
| chr14_+_93651296 | 0.10 |
ENST00000283534.4
ENST00000557574.1 |
TMEM251
RP11-371E8.4
|
transmembrane protein 251 Uncharacterized protein |
| chr3_+_101292939 | 0.09 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr19_+_57901326 | 0.09 |
ENST00000596400.1
|
AC003002.6
|
Uncharacterized protein |
| chr3_+_185000729 | 0.09 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_-_190627481 | 0.09 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr2_+_108994466 | 0.09 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr8_-_21966893 | 0.09 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr12_-_58165870 | 0.09 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr20_+_5892147 | 0.08 |
ENST00000455042.1
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr11_-_63439013 | 0.08 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr14_+_23025534 | 0.08 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr4_-_146019335 | 0.08 |
ENST00000451299.2
ENST00000507656.1 ENST00000309439.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr16_+_81040794 | 0.08 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr18_+_77867177 | 0.08 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr1_+_156030937 | 0.08 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr5_+_131892603 | 0.08 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr2_-_21022818 | 0.08 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr14_+_105219437 | 0.08 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr4_+_128802016 | 0.08 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr6_+_56819895 | 0.08 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr19_+_2841433 | 0.08 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chr10_+_134351319 | 0.08 |
ENST00000368594.3
ENST00000368593.3 |
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa |
| chr16_+_19078960 | 0.08 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr1_+_28099683 | 0.08 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr3_-_154042235 | 0.07 |
ENST00000308361.6
ENST00000496811.1 ENST00000544526.1 |
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr4_+_668348 | 0.07 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr8_-_10697281 | 0.07 |
ENST00000524114.1
ENST00000553390.1 ENST00000554914.1 |
PINX1
SOX7
SOX7
|
PIN2/TERF1 interacting, telomerase inhibitor 1 SRY (sex determining region Y)-box 7 Transcription factor SOX-7; Uncharacterized protein; cDNA FLJ58508, highly similar to Transcription factor SOX-7 |
| chr11_-_8680383 | 0.07 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr4_-_170533723 | 0.07 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr10_+_35484793 | 0.07 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr6_-_127780510 | 0.07 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr18_+_11851383 | 0.07 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chrX_-_73834449 | 0.07 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr20_-_17949100 | 0.07 |
ENST00000431277.1
|
SNX5
|
sorting nexin 5 |
| chr8_+_98788003 | 0.07 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr2_-_202645835 | 0.07 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr16_+_19079215 | 0.07 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr13_-_101327028 | 0.07 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr11_-_18343669 | 0.07 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr2_+_219745020 | 0.07 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr4_-_668108 | 0.06 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr3_-_101395936 | 0.06 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr1_+_29213584 | 0.06 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr2_+_10183651 | 0.06 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr20_-_17949363 | 0.06 |
ENST00000377759.4
ENST00000606557.1 ENST00000606602.1 ENST00000486039.1 ENST00000481323.1 |
SNX5
|
sorting nexin 5 |
| chr2_+_87769459 | 0.06 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr12_-_64616019 | 0.06 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chr11_-_31839422 | 0.06 |
ENST00000423822.2
|
PAX6
|
paired box 6 |
| chr3_-_33759541 | 0.06 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr9_+_70856899 | 0.06 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr4_-_39367949 | 0.06 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr20_-_17949143 | 0.06 |
ENST00000419004.1
|
SNX5
|
sorting nexin 5 |
| chr16_+_29911864 | 0.06 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr8_-_82754427 | 0.06 |
ENST00000353788.4
ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16
|
sorting nexin 16 |
| chr6_+_28048753 | 0.06 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr1_-_47184723 | 0.06 |
ENST00000371933.3
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr11_-_123525289 | 0.06 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr5_-_145562147 | 0.06 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr13_+_111767650 | 0.06 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr12_-_51566562 | 0.06 |
ENST00000548108.1
|
TFCP2
|
transcription factor CP2 |
| chr3_-_51937331 | 0.06 |
ENST00000310914.5
|
IQCF1
|
IQ motif containing F1 |
| chr7_+_140396946 | 0.06 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr1_+_40713573 | 0.06 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr11_+_28129795 | 0.06 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr2_+_37571717 | 0.06 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr2_+_114195268 | 0.05 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr17_-_41322332 | 0.05 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr20_+_17949544 | 0.05 |
ENST00000377710.5
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 1.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.3 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 2.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) positive regulation of fear response(GO:1903367) regulation of corticosterone secretion(GO:2000852) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0072642 | interferon-alpha secretion(GO:0072642) telomerase RNA stabilization(GO:0090669) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.3 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.2 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 2.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |