Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
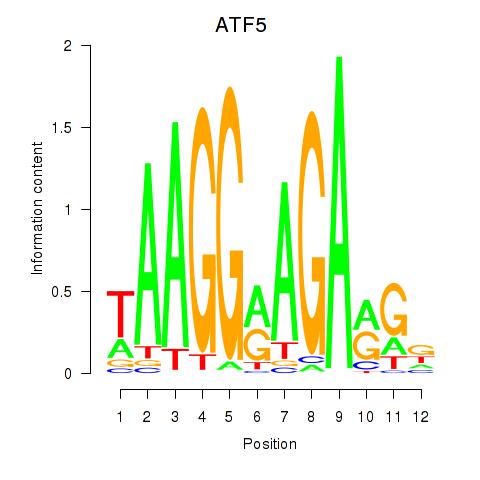
Results for ATF5
Z-value: 0.78
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50431959_50431998 | -0.49 | 3.2e-01 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_32546145 | 1.03 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr14_+_32546274 | 0.70 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_223354486 | 0.57 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr6_-_100016492 | 0.54 |
ENST00000369217.4
ENST00000369220.4 ENST00000482541.2 |
CCNC
|
cyclin C |
| chr1_+_206137237 | 0.48 |
ENST00000468509.1
ENST00000367129.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr1_+_16083098 | 0.47 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr21_-_35773370 | 0.47 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr2_-_113542063 | 0.47 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr6_-_100016678 | 0.45 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr11_+_28724129 | 0.42 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr1_+_101361626 | 0.41 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr8_+_95732095 | 0.40 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr14_+_32547434 | 0.39 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_101361782 | 0.37 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr18_-_24283586 | 0.37 |
ENST00000579458.1
|
U3
|
Small nucleolar RNA U3 |
| chr6_+_87865262 | 0.37 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr2_-_188419078 | 0.36 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chrX_+_123095546 | 0.36 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr6_-_100016527 | 0.36 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr1_+_209859510 | 0.36 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_-_88108192 | 0.35 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_-_87248975 | 0.35 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr5_-_39219705 | 0.35 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr12_-_100656134 | 0.34 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr8_+_95731904 | 0.34 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr3_-_101232019 | 0.32 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr3_-_182698381 | 0.32 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr16_-_72128270 | 0.31 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr17_+_38334501 | 0.30 |
ENST00000541245.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr2_-_188419200 | 0.30 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr22_+_37678505 | 0.30 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chr1_+_151739131 | 0.30 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr8_+_107738240 | 0.30 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr15_+_63481668 | 0.29 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr1_+_61869748 | 0.29 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr9_-_125240235 | 0.29 |
ENST00000259357.2
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1 |
| chr14_-_65569244 | 0.28 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr4_+_84377115 | 0.28 |
ENST00000295491.4
ENST00000507019.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr22_-_30662828 | 0.26 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chrX_-_122866874 | 0.26 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr6_+_31865552 | 0.25 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr5_+_175223715 | 0.25 |
ENST00000515502.1
|
CPLX2
|
complexin 2 |
| chr19_-_8642289 | 0.25 |
ENST00000596675.1
ENST00000338257.8 |
MYO1F
|
myosin IF |
| chr11_-_117102768 | 0.25 |
ENST00000532301.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr16_+_9185450 | 0.25 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr12_-_120884175 | 0.24 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr17_-_7108436 | 0.24 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr7_-_3083573 | 0.23 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr14_+_50065376 | 0.23 |
ENST00000298288.6
|
LRR1
|
leucine rich repeat protein 1 |
| chr8_-_131028869 | 0.23 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chrX_+_123095155 | 0.23 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr7_-_37488547 | 0.23 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr12_-_54778244 | 0.22 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr12_-_72057638 | 0.22 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr14_+_96968802 | 0.22 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr2_-_113999260 | 0.21 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chrX_+_131157609 | 0.21 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr11_-_7904464 | 0.21 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr3_-_23958506 | 0.21 |
ENST00000425478.2
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr14_+_21566980 | 0.21 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr9_+_131902346 | 0.21 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr4_+_84377198 | 0.20 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr14_+_96968707 | 0.20 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr18_-_812517 | 0.20 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr19_+_4229495 | 0.20 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr9_+_19230433 | 0.20 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chr4_-_84376953 | 0.20 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr5_-_39219641 | 0.20 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr3_-_69129501 | 0.20 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr2_+_234601512 | 0.19 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chrX_-_47489244 | 0.18 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr14_+_100111447 | 0.18 |
ENST00000330710.5
ENST00000357223.2 |
HHIPL1
|
HHIP-like 1 |
| chr8_+_107738343 | 0.18 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chrX_-_44402231 | 0.18 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr1_-_103574024 | 0.18 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr3_+_19988885 | 0.18 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chrX_+_123095890 | 0.17 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr5_-_148442584 | 0.16 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr19_+_15160130 | 0.16 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr17_+_7533439 | 0.16 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr3_-_23958402 | 0.16 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr17_+_33474826 | 0.15 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr2_-_209118974 | 0.15 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chrX_-_19983260 | 0.14 |
ENST00000340625.3
|
CXorf23
|
chromosome X open reading frame 23 |
| chr1_-_27286897 | 0.14 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr14_-_65569186 | 0.14 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr4_+_42399856 | 0.14 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr19_+_41509851 | 0.14 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr3_-_114173530 | 0.13 |
ENST00000470311.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chrX_+_133930798 | 0.13 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr12_-_96336369 | 0.13 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr1_+_202091980 | 0.13 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr3_-_183145873 | 0.13 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr12_+_32655048 | 0.13 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_-_104480019 | 0.13 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr18_-_812231 | 0.12 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr6_-_31514333 | 0.12 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr12_-_12491608 | 0.12 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr11_+_57310114 | 0.12 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr1_+_211433275 | 0.12 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr14_+_50065459 | 0.12 |
ENST00000318317.4
|
LRR1
|
leucine rich repeat protein 1 |
| chr8_-_21645804 | 0.12 |
ENST00000518077.1
ENST00000517892.1 |
GFRA2
|
GDNF family receptor alpha 2 |
| chr17_-_6947225 | 0.11 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr3_+_19988566 | 0.11 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr12_+_32655110 | 0.11 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr8_-_100025238 | 0.11 |
ENST00000521696.1
|
RP11-410L14.2
|
RP11-410L14.2 |
| chr5_-_148442687 | 0.11 |
ENST00000515425.1
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr5_+_148786518 | 0.11 |
ENST00000518014.1
ENST00000505340.1 ENST00000509909.1 |
AC131025.8
MIR143HG
|
AC131025.8 MIR143 host gene (non-protein coding) |
| chr5_-_132200477 | 0.11 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr3_-_88108212 | 0.11 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr14_+_79746249 | 0.10 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr2_+_153191706 | 0.10 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr12_+_10124001 | 0.10 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr6_+_76599809 | 0.10 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr17_+_33474860 | 0.10 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr5_-_20575959 | 0.10 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr17_+_33914424 | 0.10 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr19_-_18366182 | 0.10 |
ENST00000355502.3
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific |
| chr12_-_1920886 | 0.09 |
ENST00000536846.2
ENST00000538027.2 ENST00000538450.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr21_-_28215332 | 0.09 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_+_23958470 | 0.09 |
ENST00000434031.2
ENST00000413699.1 ENST00000456530.2 |
RPL15
|
ribosomal protein L15 |
| chr5_+_82767487 | 0.09 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr2_-_225434538 | 0.09 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr3_-_187454281 | 0.09 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr2_-_145278475 | 0.09 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_3607228 | 0.08 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr5_+_154393260 | 0.08 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr7_-_78400598 | 0.08 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_208994548 | 0.08 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr5_+_50678921 | 0.08 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr1_-_47697387 | 0.08 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr7_-_127672146 | 0.08 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr2_-_121624973 | 0.08 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chrX_+_123095860 | 0.08 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr3_+_190105909 | 0.08 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr5_-_180552304 | 0.08 |
ENST00000329365.2
|
OR2V1
|
olfactory receptor, family 2, subfamily V, member 1 |
| chr7_+_150688166 | 0.08 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr12_-_16761117 | 0.08 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_+_2800722 | 0.07 |
ENST00000508385.1
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr3_+_112930387 | 0.07 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr2_-_197036289 | 0.07 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chrX_+_130677950 | 0.07 |
ENST00000338616.3
|
OR13H1
|
olfactory receptor, family 13, subfamily H, member 1 |
| chr4_-_68620053 | 0.07 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr5_+_132009675 | 0.07 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr15_-_90222642 | 0.07 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr8_+_67344710 | 0.07 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr17_+_47287749 | 0.07 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr2_-_87018784 | 0.07 |
ENST00000283635.3
ENST00000538832.1 |
CD8A
|
CD8a molecule |
| chr4_+_48807155 | 0.06 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr9_-_6007787 | 0.06 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr8_-_21646290 | 0.06 |
ENST00000519195.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr19_+_55795493 | 0.06 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr19_-_7936344 | 0.06 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chrX_+_153409678 | 0.06 |
ENST00000369951.4
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive |
| chr13_-_36871886 | 0.06 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr16_-_72206034 | 0.05 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr2_+_149447783 | 0.05 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr18_-_53089538 | 0.05 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr5_+_82767583 | 0.05 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr1_-_17304771 | 0.05 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chr1_-_22109484 | 0.05 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr9_+_2621798 | 0.05 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr5_+_82767284 | 0.05 |
ENST00000265077.3
|
VCAN
|
versican |
| chr1_-_22109682 | 0.05 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr7_+_143771275 | 0.05 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr10_+_111765562 | 0.05 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr5_+_138629417 | 0.05 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr1_-_183387723 | 0.04 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr15_+_75118888 | 0.04 |
ENST00000395018.4
|
CPLX3
|
complexin 3 |
| chr11_-_106889250 | 0.04 |
ENST00000526355.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chr16_-_65155979 | 0.04 |
ENST00000562325.1
ENST00000268603.4 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr16_+_28943260 | 0.04 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr14_-_65569057 | 0.04 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr22_-_37215523 | 0.04 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr18_-_53089723 | 0.04 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr5_+_110559603 | 0.04 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_+_23958632 | 0.04 |
ENST00000412097.1
ENST00000415719.1 ENST00000435882.1 |
RPL15
|
ribosomal protein L15 |
| chr10_+_103113840 | 0.04 |
ENST00000393441.4
ENST00000408038.2 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr17_+_44668387 | 0.04 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chrX_+_122318006 | 0.04 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr3_-_183145765 | 0.04 |
ENST00000473233.1
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr7_+_128399002 | 0.04 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr16_+_22103828 | 0.04 |
ENST00000567131.1
ENST00000568328.1 ENST00000389398.5 ENST00000389397.4 |
VWA3A
|
von Willebrand factor A domain containing 3A |
| chr1_+_48688357 | 0.04 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr9_+_2621950 | 0.03 |
ENST00000382096.1
|
VLDLR
|
very low density lipoprotein receptor |
| chr19_-_38916822 | 0.03 |
ENST00000586305.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr5_+_50679506 | 0.03 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr7_-_27142290 | 0.03 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr1_+_16083123 | 0.03 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr8_-_95449155 | 0.03 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr2_+_88047606 | 0.03 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr4_-_175750364 | 0.03 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr3_-_62861012 | 0.03 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr3_-_114173654 | 0.03 |
ENST00000482689.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_78400364 | 0.03 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_79745746 | 0.03 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr16_-_65155833 | 0.03 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr14_+_92789498 | 0.03 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr8_-_95220775 | 0.03 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr2_-_209119831 | 0.02 |
ENST00000345146.2
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.7 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.8 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.3 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.2 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0071288 | B cell costimulation(GO:0031296) cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.0 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |