Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
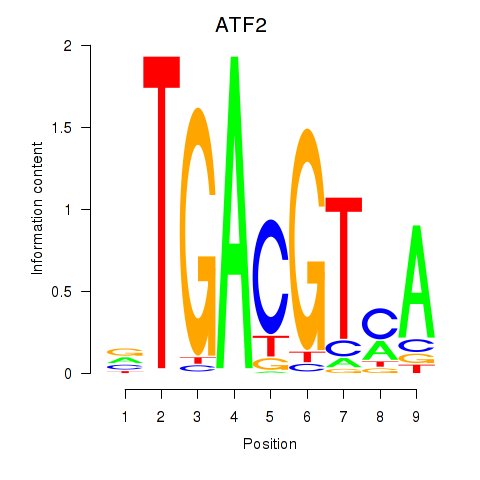
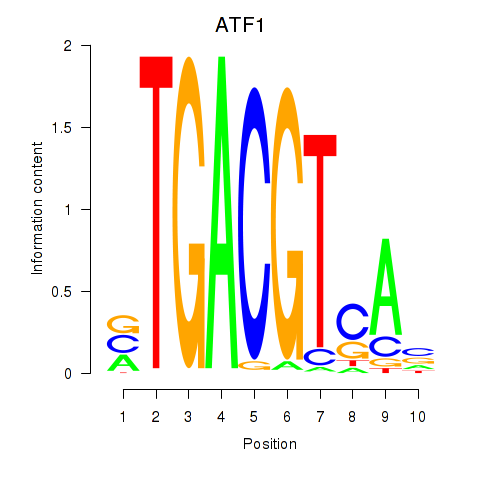
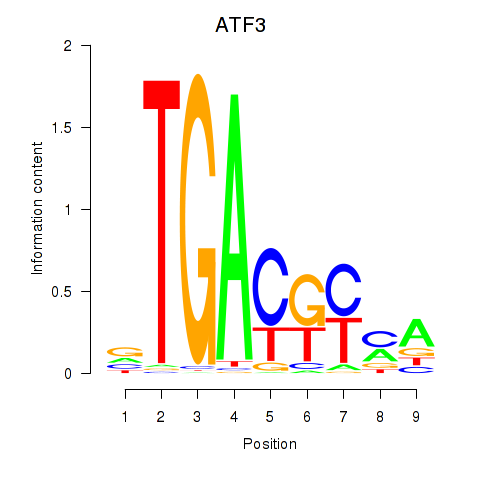
Results for ATF2_ATF1_ATF3
Z-value: 0.37
Transcription factors associated with ATF2_ATF1_ATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF2
|
ENSG00000115966.12 | activating transcription factor 2 |
|
ATF1
|
ENSG00000123268.4 | activating transcription factor 1 |
|
ATF3
|
ENSG00000162772.12 | activating transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF3 | hg19_v2_chr1_+_212738676_212738755 | 0.77 | 7.4e-02 | Click! |
| ATF2 | hg19_v2_chr2_-_176032843_176032941 | -0.45 | 3.7e-01 | Click! |
| ATF1 | hg19_v2_chr12_+_51158263_51158395 | -0.38 | 4.6e-01 | Click! |
Activity profile of ATF2_ATF1_ATF3 motif
Sorted Z-values of ATF2_ATF1_ATF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33292507 | 0.29 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr6_-_38607628 | 0.26 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr1_+_156698743 | 0.26 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr19_+_3762703 | 0.25 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr6_-_31940065 | 0.22 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr16_+_19078960 | 0.22 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr7_+_30174574 | 0.21 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr8_+_103876528 | 0.20 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr13_+_111972980 | 0.19 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr12_-_123380610 | 0.19 |
ENST00000535765.1
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr14_+_93389425 | 0.18 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr19_+_50979753 | 0.17 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr6_-_121655850 | 0.17 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr12_+_10365082 | 0.17 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_+_9066472 | 0.16 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr1_-_713985 | 0.16 |
ENST00000428504.1
|
RP11-206L10.2
|
RP11-206L10.2 |
| chr19_+_36024310 | 0.16 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr5_-_99870932 | 0.15 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr8_-_21966893 | 0.15 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr20_+_33146510 | 0.15 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr19_-_46088068 | 0.15 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr10_+_35484793 | 0.14 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr17_+_48585902 | 0.14 |
ENST00000452039.1
|
MYCBPAP
|
MYCBP associated protein |
| chr7_+_139025875 | 0.14 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr13_-_33002151 | 0.14 |
ENST00000495479.1
ENST00000343281.4 ENST00000464470.1 ENST00000380139.4 ENST00000380133.2 |
N4BP2L1
|
NEDD4 binding protein 2-like 1 |
| chr13_-_52378231 | 0.14 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr1_+_228645796 | 0.14 |
ENST00000369160.2
|
HIST3H2BB
|
histone cluster 3, H2bb |
| chr7_+_30174668 | 0.13 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr17_-_8059638 | 0.13 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr6_-_34664612 | 0.13 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr3_-_12587055 | 0.13 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr6_-_31939734 | 0.12 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr22_-_39097527 | 0.12 |
ENST00000417712.1
|
JOSD1
|
Josephin domain containing 1 |
| chr11_+_66610883 | 0.12 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr2_-_74781061 | 0.12 |
ENST00000264094.3
ENST00000393937.2 ENST00000409986.1 |
LOXL3
|
lysyl oxidase-like 3 |
| chr17_+_48585794 | 0.12 |
ENST00000576179.1
ENST00000419930.1 |
MYCBPAP
|
MYCBP associated protein |
| chr12_+_14956506 | 0.12 |
ENST00000330828.2
|
C12orf60
|
chromosome 12 open reading frame 60 |
| chr19_+_54960790 | 0.11 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr1_-_220608023 | 0.11 |
ENST00000416839.2
|
AC096644.1
|
Uncharacterized protein |
| chr6_-_41888843 | 0.11 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr1_-_119682812 | 0.11 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr8_+_86999516 | 0.11 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr19_-_44100275 | 0.11 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr16_+_2564254 | 0.11 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr17_+_33448593 | 0.11 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr16_-_3068171 | 0.11 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr6_-_47445214 | 0.11 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr1_+_154909803 | 0.11 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr6_+_11537910 | 0.11 |
ENST00000543875.1
|
TMEM170B
|
transmembrane protein 170B |
| chrX_-_106960285 | 0.10 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr7_-_45026159 | 0.10 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr19_+_13858701 | 0.10 |
ENST00000586666.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr21_-_44299626 | 0.10 |
ENST00000330317.2
ENST00000398208.2 |
WDR4
|
WD repeat domain 4 |
| chr19_+_45971246 | 0.10 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr1_-_151254362 | 0.10 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr16_+_29911666 | 0.10 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr4_+_119512928 | 0.10 |
ENST00000567913.2
|
RP11-384K6.6
|
RP11-384K6.6 |
| chr16_+_67927147 | 0.10 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr8_+_23386557 | 0.10 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr3_+_128598433 | 0.10 |
ENST00000308982.7
ENST00000514336.1 |
ACAD9
|
acyl-CoA dehydrogenase family, member 9 |
| chr19_-_18314799 | 0.10 |
ENST00000481914.2
|
RAB3A
|
RAB3A, member RAS oncogene family |
| chr1_+_156698708 | 0.10 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr6_-_38607673 | 0.10 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr15_-_72612470 | 0.10 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr1_+_1217489 | 0.10 |
ENST00000325425.8
ENST00000400928.3 |
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chrX_+_48432892 | 0.09 |
ENST00000376759.3
ENST00000430348.2 |
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr6_-_27782548 | 0.09 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr13_-_36871886 | 0.09 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr12_+_10365404 | 0.09 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr7_+_30174426 | 0.09 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr11_-_66056596 | 0.09 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr15_-_32162833 | 0.09 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr17_+_7155343 | 0.09 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr16_+_31044413 | 0.09 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr3_-_134092561 | 0.09 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr17_+_7155556 | 0.09 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr6_-_116575226 | 0.09 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr17_+_7211656 | 0.08 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr10_+_134210672 | 0.08 |
ENST00000305233.5
ENST00000368609.4 |
PWWP2B
|
PWWP domain containing 2B |
| chr11_+_12696102 | 0.08 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr2_-_98280383 | 0.08 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr16_-_86588627 | 0.08 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chr21_+_43619796 | 0.08 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr11_-_66056478 | 0.08 |
ENST00000431556.2
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr14_+_63671105 | 0.08 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chrX_+_155227246 | 0.08 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr1_-_161147275 | 0.08 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr16_+_3068393 | 0.08 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr7_-_45026200 | 0.08 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr17_-_76836963 | 0.08 |
ENST00000312010.6
|
USP36
|
ubiquitin specific peptidase 36 |
| chr17_-_47841485 | 0.08 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr8_-_82754193 | 0.08 |
ENST00000519817.1
ENST00000521773.1 ENST00000523757.1 |
SNX16
|
sorting nexin 16 |
| chr19_-_55574538 | 0.08 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr13_-_52378281 | 0.08 |
ENST00000218981.1
|
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr6_+_143381979 | 0.08 |
ENST00000367598.5
ENST00000447498.1 ENST00000357847.4 ENST00000344492.5 ENST00000367596.1 ENST00000494282.2 ENST00000275235.4 |
AIG1
|
androgen-induced 1 |
| chr9_-_100000957 | 0.08 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr5_-_175964366 | 0.08 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr19_-_2944907 | 0.08 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr19_-_47616992 | 0.08 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr19_+_57874835 | 0.08 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr14_+_91526668 | 0.08 |
ENST00000521334.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr2_+_70142232 | 0.08 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr17_-_77967433 | 0.08 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr8_-_95274536 | 0.08 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr7_-_6388537 | 0.08 |
ENST00000313324.4
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220, member A |
| chr16_+_29911864 | 0.08 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr14_+_102276192 | 0.08 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr17_+_7835419 | 0.08 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr7_-_45151272 | 0.07 |
ENST00000461363.1
ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr12_-_371994 | 0.07 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr19_+_13858593 | 0.07 |
ENST00000221554.8
|
CCDC130
|
coiled-coil domain containing 130 |
| chr1_-_17231271 | 0.07 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr11_+_3819049 | 0.07 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr2_-_220110111 | 0.07 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chrX_+_49832231 | 0.07 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr17_-_15902903 | 0.07 |
ENST00000486655.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr19_-_50979981 | 0.07 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr17_+_7155819 | 0.07 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr3_+_150126101 | 0.07 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr20_+_33292068 | 0.07 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_+_27071002 | 0.07 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr19_-_51014345 | 0.07 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr8_-_102803163 | 0.07 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr12_+_133707570 | 0.07 |
ENST00000416488.1
ENST00000540096.2 |
ZNF268
CTD-2140B24.4
|
zinc finger protein 268 Zinc finger protein 268 |
| chr19_+_36103631 | 0.07 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr5_+_172332220 | 0.07 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr16_+_20911978 | 0.07 |
ENST00000562740.1
|
LYRM1
|
LYR motif containing 1 |
| chr17_-_43210580 | 0.07 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr11_-_9482010 | 0.07 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr11_+_69455855 | 0.07 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr17_-_76836729 | 0.07 |
ENST00000587783.1
ENST00000542802.3 ENST00000586531.1 ENST00000589424.1 ENST00000590546.2 |
USP36
|
ubiquitin specific peptidase 36 |
| chr6_-_3157760 | 0.07 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr4_+_667686 | 0.07 |
ENST00000505477.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr11_-_64163297 | 0.07 |
ENST00000457725.1
|
AP003774.6
|
AP003774.6 |
| chr22_-_19132154 | 0.07 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr16_+_70380825 | 0.07 |
ENST00000417604.2
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chr6_+_30029008 | 0.07 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr10_+_99496872 | 0.07 |
ENST00000337540.7
ENST00000357540.4 ENST00000370613.3 ENST00000370610.3 ENST00000393677.4 ENST00000453958.2 ENST00000359980.3 |
ZFYVE27
|
zinc finger, FYVE domain containing 27 |
| chr6_+_26204825 | 0.07 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr11_-_14993819 | 0.07 |
ENST00000396372.2
ENST00000361010.3 ENST00000359642.3 ENST00000331587.4 |
CALCA
|
calcitonin-related polypeptide alpha |
| chr19_-_50528392 | 0.07 |
ENST00000600137.1
ENST00000597215.1 |
VRK3
|
vaccinia related kinase 3 |
| chr17_-_27038063 | 0.07 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr19_-_40919271 | 0.07 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr7_-_129592700 | 0.07 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr19_-_18314836 | 0.07 |
ENST00000464076.3
ENST00000222256.4 |
RAB3A
|
RAB3A, member RAS oncogene family |
| chr1_-_154909329 | 0.07 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr17_+_43209967 | 0.07 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr11_-_95523500 | 0.07 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr1_-_32110467 | 0.06 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr16_+_27561449 | 0.06 |
ENST00000261588.4
|
KIAA0556
|
KIAA0556 |
| chr21_-_43916433 | 0.06 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr11_-_114271139 | 0.06 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr19_+_54704610 | 0.06 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr1_-_119683251 | 0.06 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr5_-_132948216 | 0.06 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chrX_+_155227371 | 0.06 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
| chr3_+_16306837 | 0.06 |
ENST00000606098.1
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr18_-_73967160 | 0.06 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr6_+_43027595 | 0.06 |
ENST00000259708.3
ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4
|
kinesin light chain 4 |
| chr17_-_685493 | 0.06 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr2_-_220408260 | 0.06 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chrX_+_152338301 | 0.06 |
ENST00000453825.2
|
PNMA6A
|
paraneoplastic Ma antigen family member 6A |
| chr6_-_31671058 | 0.06 |
ENST00000538874.1
ENST00000395952.3 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr1_-_26700943 | 0.06 |
ENST00000416125.1
|
ZNF683
|
zinc finger protein 683 |
| chr1_+_45265897 | 0.06 |
ENST00000372201.4
|
PLK3
|
polo-like kinase 3 |
| chr1_+_149239529 | 0.06 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr19_+_49375649 | 0.06 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr16_-_67881588 | 0.06 |
ENST00000561593.1
ENST00000565114.1 |
CENPT
|
centromere protein T |
| chr19_-_52227221 | 0.06 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr16_+_70380732 | 0.06 |
ENST00000302243.7
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chr17_-_76778339 | 0.06 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr19_+_19030497 | 0.06 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr7_+_23719749 | 0.06 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr5_-_147211226 | 0.06 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr16_+_30087288 | 0.06 |
ENST00000279387.7
ENST00000562664.1 ENST00000562222.1 |
PPP4C
|
protein phosphatase 4, catalytic subunit |
| chr1_+_16083098 | 0.06 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_+_152240819 | 0.06 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr5_-_99870890 | 0.06 |
ENST00000499025.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr9_-_99381660 | 0.06 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr2_-_224467093 | 0.06 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr19_+_54705025 | 0.06 |
ENST00000441429.1
|
RPS9
|
ribosomal protein S9 |
| chr11_+_117049445 | 0.06 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr16_-_2318373 | 0.06 |
ENST00000566458.1
ENST00000320225.5 |
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr19_+_54704990 | 0.06 |
ENST00000391753.2
|
RPS9
|
ribosomal protein S9 |
| chr17_-_80231557 | 0.06 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr7_-_38407770 | 0.06 |
ENST00000390348.2
|
TRGV1
|
T cell receptor gamma variable 1 (non-functional) |
| chr20_+_5892037 | 0.06 |
ENST00000378961.4
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr2_-_220110187 | 0.06 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr2_+_110656268 | 0.06 |
ENST00000553749.1
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chrX_-_152245978 | 0.06 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr11_-_67276100 | 0.06 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr19_-_8070474 | 0.06 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr20_-_62258394 | 0.06 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr22_-_36850991 | 0.06 |
ENST00000442579.1
|
RP5-1119A7.14
|
RP5-1119A7.14 |
| chr19_+_42580274 | 0.06 |
ENST00000359044.4
|
ZNF574
|
zinc finger protein 574 |
| chr17_+_55333876 | 0.06 |
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2 |
| chr2_+_30454390 | 0.06 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr17_+_71161140 | 0.06 |
ENST00000357585.2
|
SSTR2
|
somatostatin receptor 2 |
| chr12_+_122150646 | 0.06 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr2_-_219524193 | 0.06 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF2_ATF1_ATF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.2 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.2 | GO:1990709 | maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.1 | 0.4 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.1 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.3 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.0 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:1903382 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.0 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.0 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.0 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.0 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.0 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.0 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |